Analysis of Single Cell RNA Sequencing Data to Define Biomarkers of Human Liver Immune Tolerance
1Surgery, University of Southern California Keck School of Medicine, Los Angeles, CA, 2Molecular Microbiology and Immunology, University of Southern California, Los Angeles, CA, 3Norris Medical Library, University of Southern California Keck School of Medicine, Los Angeles, CA, 4Ellison Institute, University of Southern California, Los Angeles, CA
Meeting: 2021 American Transplant Congress
Abstract number: 670
Keywords: Gene expression, Genomic markers, Liver transplantation, Tolerance
Topic: Clinical Science » Biomarkers, Immune Assessment and Clinical Outcomes
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes
Session Type: Poster Abstract
Session Date & Time: None. Available on demand.
Location: Virtual
*Purpose: The liver is unique in its ability to maintain immune homeostasis and promote operational tolerance following solid organ transplantation. Single-cell RNA sequencing (scRNA seq) is a powerful approach to generate highly dimensional transcriptome data to understand cellular phenotypes. However, when scRNA data is produced by different groups, with different data models, different standards, and samples processed in different ways, it can be extremely challenging to draw meaningful conclusions from the aggregated data. The goal of this study was to establish a method to combine ‘human liver’ scRNA seq datasets by 1. characterizing the heterogeneity between studies and 2. using the meta-dataset to define the immunotolerant phenotypes across immune cell subpopulations in healthy human liver.
*Methods: Publicly available scRNA seq data generated from liver samples obtained from a combined total of 17 patients were analyzed. Liver-specific immune cells (CD45+) were extracted from each dataset, and immune cell subpopulations (myeloid cells, NK and T cells, plasma cells, and B cells) were examined using dimensionality reduction (UMAP), differential gene expression, and ingenuity pathway analysis.
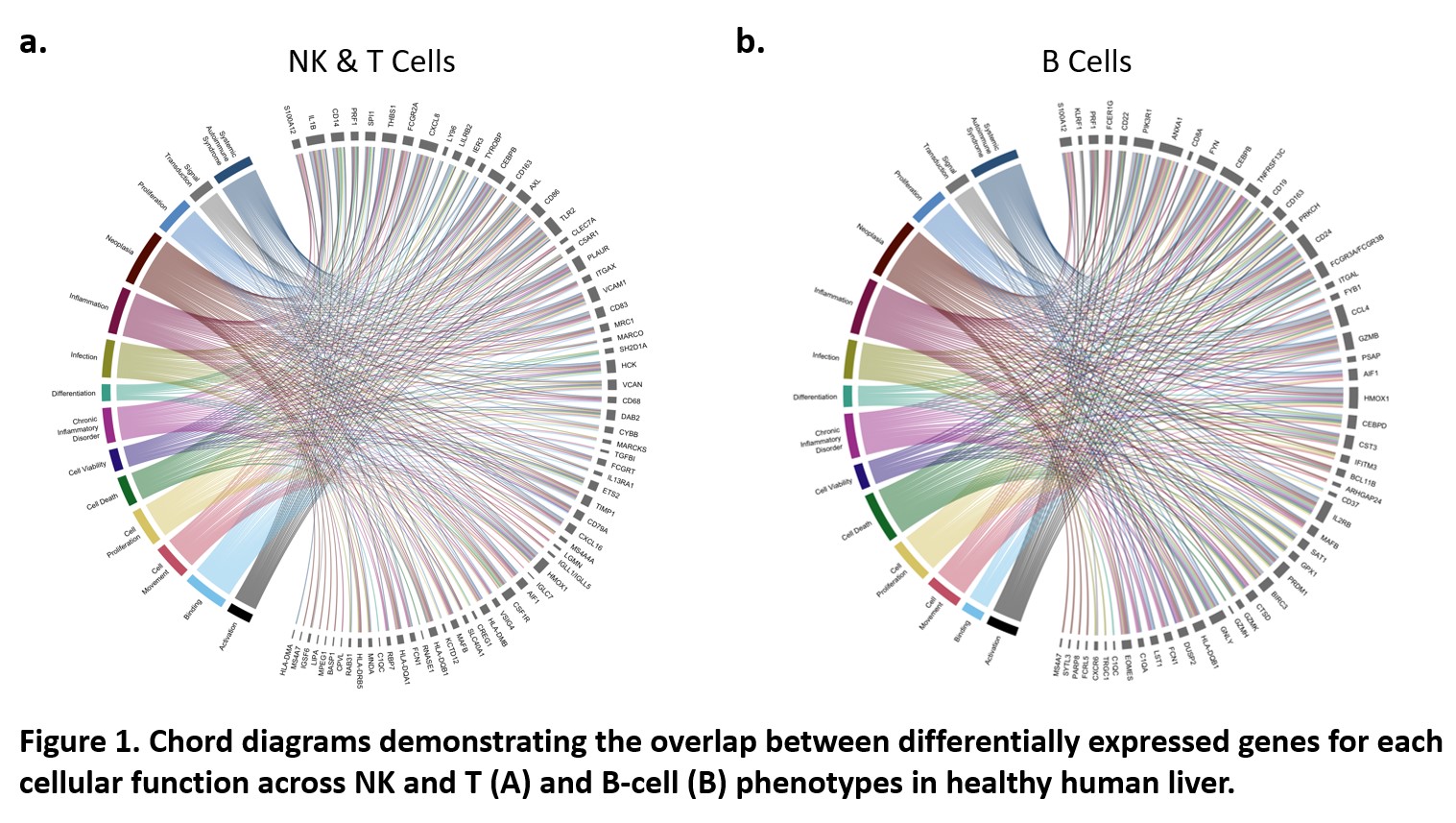
*Results: All datasets co-clustered, but cell proportions differed across studies. Gene expression correlation demonstrated similarity across all studies, and canonical pathways that differed between datasets were related to cell stress and oxidative phosphorylation rather than immune-related function. Detailed analysis of differential gene expression identified concordant gene signatures for each hepatic immune subpopulation (Fig 1). Expression levels across all immune cellular functions including immune response, activation, phagocytosis and adhesion were globally low indicating a tolerogenic state.
*Conclusions: This method for meta-analysis of scRNA sequencing datasets provides a novel approach to define the features of immune homeostasis in human liver. Pathways and cellular phenotypes involved in liver immune cell tolerance provide a critical reference point as clinical tolerance strategies are explored in liver transplant recipients.
To cite this abstract in AMA style:
Rocque B, Barbetta A, Singh P, Goldbeck C, Helou DG, Loh E, Ung N, Lee JS, Akbari O, Emamaullee J. Analysis of Single Cell RNA Sequencing Data to Define Biomarkers of Human Liver Immune Tolerance [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/analysis-of-single-cell-rna-sequencing-data-to-define-biomarkers-of-human-liver-immune-tolerance/. Accessed February 16, 2026.« Back to 2021 American Transplant Congress