Early Detection of HCC Mutations in Indeterminate Lesions Using Liquid Biopsies
1Washington University, St. Louis, MO
2Baylor University, Dallas, TX
3Stanford University, Palo Alto, CA.
Meeting: 2015 American Transplant Congress
Abstract number: C284
Keywords: Biopsy, Genomic markers, Hepatocellular carcinoma, Malignancy
Session Information
Session Name: Poster Session C: Translational Biomarkers and Immune Monitoring
Session Type: Poster Session
Date: Monday, May 4, 2015
Session Time: 5:30pm-6:30pm
 Presentation Time: 5:30pm-6:30pm
Presentation Time: 5:30pm-6:30pm
Location: Exhibit Hall E
Background Recent sequencing efforts have characterized the mutation profile of hepatocellular carcinoma (HCC); however, the relevance to clinical decision-making has been limited by the infrequency of tissue biopsies in practice. Performing a “liquid biopsy” by analyzing the cell-free DNA (cfDNA) extracted from the plasma may allow for the assessment of tumor mutations without tissue. The purpose of this study was to detect HCC mutations in cfDNA by using next-generation sequencing of six genes commonly mutated in HCC: TP53, CTNNB1, ARID1A, AXIN1, CDKN2A and TERT.
Methods cfDNA from the plasma of cirrhotic subjects with HCC were used to create sequencing libraries. We captured the exonic DNA from the six targeted genes and performed paired-end 100bp sequencing on Illumina MiSeq. We then identified mutations which were non-synonymous and not variants in the normal population. The ratio of reads containing such mutations provided a normalized score that we hypothesize to be proportional to tumor burden.
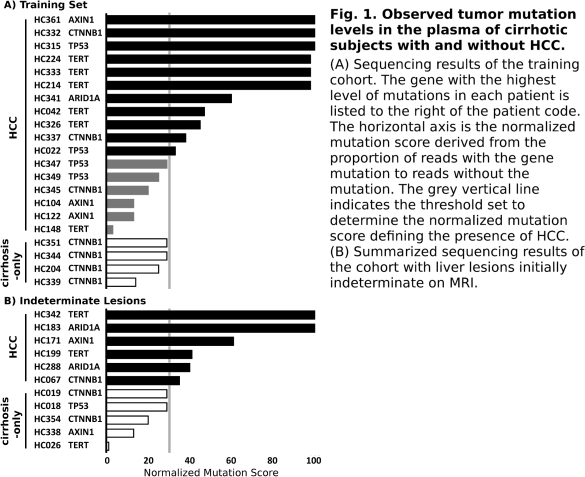
Results In our training set, we sequenced the cfDNA from 17 cirrhotic subjects with OPTN class 5 radiographic lesions; 4 samples from subjects with cirrhosis but no HCC served as controls. To identify HCC samples with maximum specificity (ie no false positives), we set the mutation score threshold to be the highest observed mutation level in the control group. This threshold categorized 11 of the 17 samples with HCC (64.6%) as having detectable HCC mutations in cfDNA (Fig. 1)—a level consistent with results from large-scale HCC sequencing studies.
To further evaluate, cfDNA was sequenced from 11 cirrhotic subjects all with indeterminate liver lesions on MRI. We predicted that 6 of these patients had early HCC, and upon follow-up, all were confirmed to have HCC by surgical pathology or subsequent imaging. The 5 subjects not predicted to have HCC remained cancer-free at a mean of 19 months of follow-up (p<0.005, Fisher's). Ongoing sequencing of larger cohorts will define optimal genes and thresholds for sequencing to better evaluate and treat HCC.
To cite this abstract in AMA style:
Lin Y, Cavaness K, Zhang Z, Lin S, Korenblat K, Klintmalm G, Chapman W. Early Detection of HCC Mutations in Indeterminate Lesions Using Liquid Biopsies [abstract]. Am J Transplant. 2015; 15 (suppl 3). https://atcmeetingabstracts.com/abstract/early-detection-of-hcc-mutations-in-indeterminate-lesions-using-liquid-biopsies/. Accessed March 2, 2026.« Back to 2015 American Transplant Congress
