Differentially Expressed Genes in Immunosuppressed Kidney Transplant Recipients
1Medicine, MMRF, HCMC, Minneapolis, MN
2Biostatistics, University of MN, Minneapolis, MN
3Pharmacy, University of MN, Minneapolis, MN
4Surgery, University of MN, Minneapolis, MN
5Medicine, University of PA, Philadelphia, PA.
Meeting: 2015 American Transplant Congress
Abstract number: B260
Keywords: Gene expression, Genomics, Kidney transplantation, T cell receptors (TcR)
Session Information
Session Name: Poster Session B: Translational Genetics and Proteomics in Transplantation
Session Type: Poster Session
Date: Sunday, May 3, 2015
Session Time: 5:30pm-6:30pm
 Presentation Time: 5:30pm-6:30pm
Presentation Time: 5:30pm-6:30pm
Location: Exhibit Hall E
Introduction: In our effort to personalize immunosuppressive therapies we performed RNA sequencing (RNAseq) to identify differentially expressed genes (DEGs) after kidney transplant (tx) and the start of immunosuppressive drugs. RNAseq is superior to microarray to determine DEGs because it's not limited to available probes, increased sensitivity, and detects alternative and previously unknown transcripts.
Methods: DEGs were determined in 42 adult tx recipients receiving antibody induction, calcineurin inhibition, mycophenolate, with and without steroids. Blood was obtained pre-tx (baseline), week 1, months 3 and 6 post-tx. PBMCs were isolated, RNA extracted and gene expression measured using RNAseq. Principal components (PCs) were computed using a surrogate variable approach. Expression was analyzed using a linear mixed effects model adjusting for age, gender and the first two PCs. DEGs post-tx were identified by controlling false discovery rate (FDR) at <0.01. DEGs with 2 fold change were assessed with Ingenuity Pathway Analysis.
Results: Week 1 and month 3 had similar DEGs.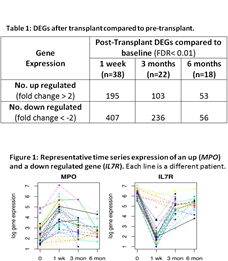 Top 10 up regulated DEGs (fold change at week 1): OLFM4 (17.5), NREP (11.7), LTF (10.7), LOXL3 (9.7), SIAE (9.4), MPO (9.0), PRTN3 (8.0), LCN2 (7.4), OLR1 (7.3), UQCC2 (7.0). Top 10 down regulated DEGs at week 1: IL7R (-24.5), CD3E (-19.5), KLRB1 (-19.3), CD3D (-18.8), LEF1 (-16.9), ITK (-16.5), CD3G (-16.2), GZMK (-15.4), CD2 (-15.0), MAL (-14.6). DEGs at 6 months post-tx involve a different gene signature.
Top 10 up regulated DEGs (fold change at week 1): OLFM4 (17.5), NREP (11.7), LTF (10.7), LOXL3 (9.7), SIAE (9.4), MPO (9.0), PRTN3 (8.0), LCN2 (7.4), OLR1 (7.3), UQCC2 (7.0). Top 10 down regulated DEGs at week 1: IL7R (-24.5), CD3E (-19.5), KLRB1 (-19.3), CD3D (-18.8), LEF1 (-16.9), ITK (-16.5), CD3G (-16.2), GZMK (-15.4), CD2 (-15.0), MAL (-14.6). DEGs at 6 months post-tx involve a different gene signature.
Conclusions: More DEGs were down than up regulated and the number of DEGs decreases with time post-tx. Top altered pathways at 1 week and 3 month post-tx are: iCOS-iCOSL signaling, T helper cell differentiation and CD28 signaling, T cell receptor signaling and CTLA4 signaling in CD8 T Cells. (Funded by NIGMS)
To cite this abstract in AMA style:
Dorr C, Wu B, Guan W, Muthusamy A, Sanghavi K, Jacobson P, Oetting W, Schladt D, Remmel R, Maltzman J, Matas A, Investigators ForDeKAFGenomics, Israni A. Differentially Expressed Genes in Immunosuppressed Kidney Transplant Recipients [abstract]. Am J Transplant. 2015; 15 (suppl 3). https://atcmeetingabstracts.com/abstract/differentially-expressed-genes-in-immunosuppressed-kidney-transplant-recipients/. Accessed March 9, 2026.« Back to 2015 American Transplant Congress
