Comprehensive and Combined Transcriptomic and Proteomic Analysis Reveals Key Factors Underlying Ischemia-Reperfusion Injury in Human Liver Transplantation
Organ Transplant Center, the First Affiliated Hospital of SYSU, Guangzhou, China
Meeting: 2019 American Transplant Congress
Abstract number: D260
Keywords: Gene expression, Genomics, Ischemia, Liver transplantation
Session Information
Session Name: Poster Session D: Non-Organ Specific:Organ Preservation/Ischemia Reperfusion Injury
Session Type: Poster Session
Date: Tuesday, June 4, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
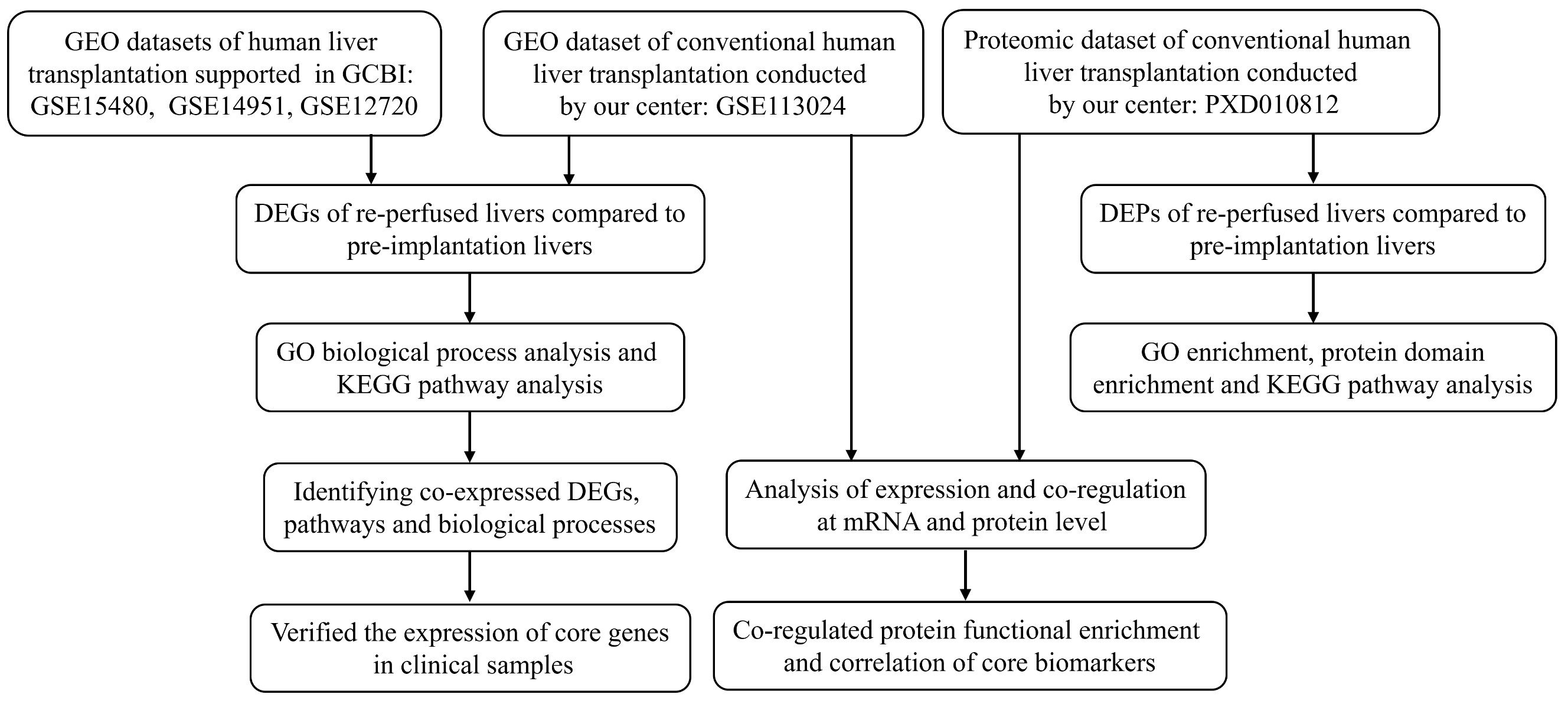
*Purpose: Ischemia-reperfusion injury (IRI) is a major challenge in liver resection and liver transplantation surgery. Hepatic IRI can seriously impair liver function and even cause irreversible damage, but there are gaps in the literature regarding IRI. Herein, we aimed to explore the possible molecular mechanisms underlying IRI using comprehensive and combined bioinformatics analyses.
*Methods: Three Gene Expression Omnibus (GEO) datasets comprising liver transplantation data (GSE15480, GSE14951, and GSE12720) and a dataset constructed by our center (GSE113024) were collected for a comprehensive analysis. The co-regulated differentially expressed genes (DEGs), intersecting pathways and key biological processes were identified and validated in clinical samples. We then performed a proteomic analysis of ischemic and reperfused livers (PXD010812). Differentially expressed proteins, functional enrichment involving GO terms, protein domain enrichment and KEGG pathways were identified. Correlations between the transcriptome and proteome were analyzed, and co-regulated DEGs and co-regulated markers were used to elucidate the core factors underlying IRI.
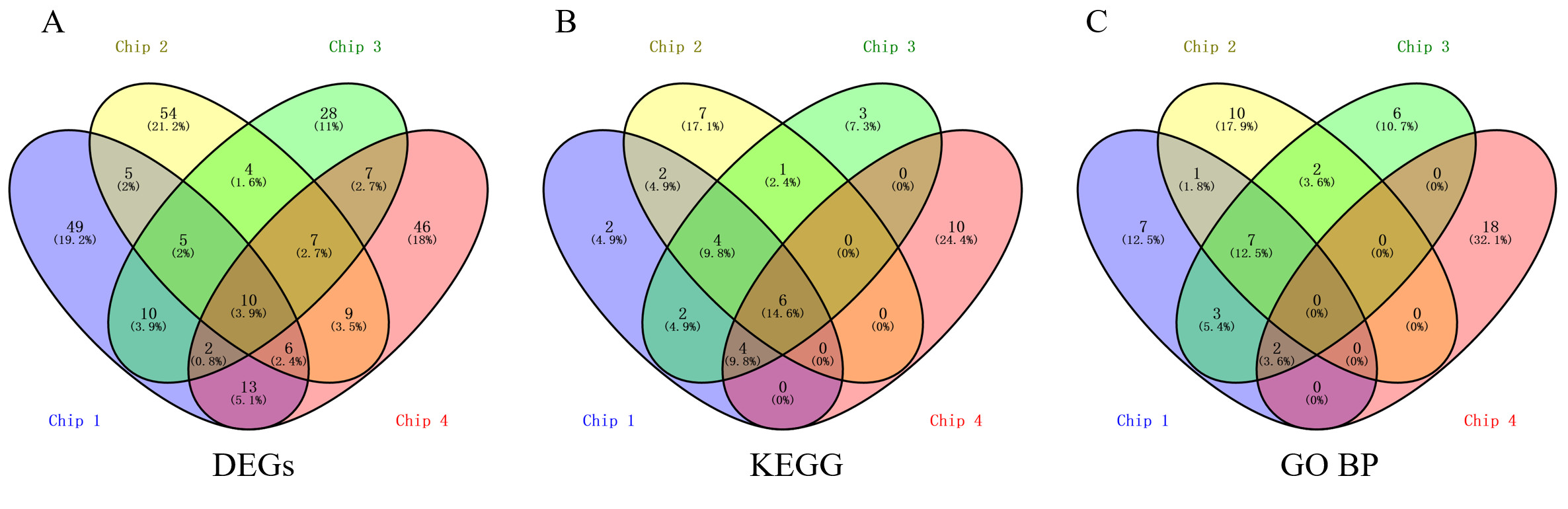
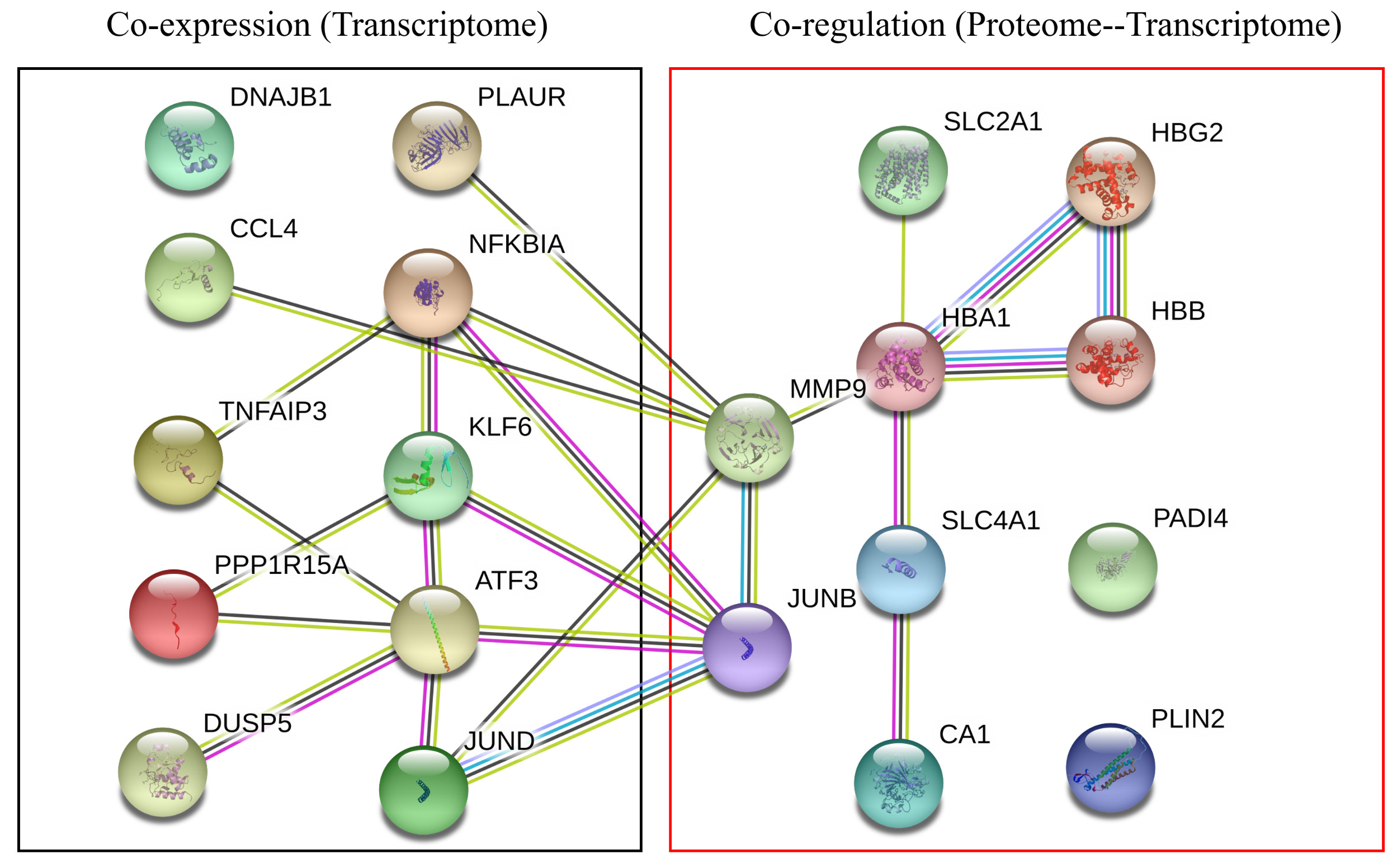
*Results: As shown in Figure below, ten DEGs were co-upregulated in the 4 GEO datasets, including ATF3, CCL4, DNAJB1, DUSP5, JUND, KLF6, NFKBIA, PLAUR, PPP1R15A, and TNFAIP3. Co-expression of the DEGs in 10 matched clinical samples was verified by quantitative real-time PCR.
Six pathways were co-enriched in all 4 chips, including the NF-kappa B signaling pathway; NOD-like receptor signaling pathway; Influenza A; Legionellosis; Transcriptional misregulation in cancer; and MAPK signaling pathway. The combined analysis demonstrated 10 co-regulated genes/proteins, including HBB, HBG2, CA1, SLC4A1, PLIN2, JUNB, HBA1, MMP9, SLC2A1, and PADI4. The co-regulated DEGs and co-regulated genes/proteins formed a tight interaction network and could serve as the core factors underlying IRI .
*Conclusions: Comprehensive and combined transcriptomic and proteomic analyses revealed key factors underlying IRI during human liver transplantation. Further studies are needed to elucidate the detailed mechanisms of these potential key factors in IRI progression.
To cite this abstract in AMA style:
Huang S, He X, Sun C, Guo Z, Ju W, Zhu Z, Zhao Q, Chen G. Comprehensive and Combined Transcriptomic and Proteomic Analysis Reveals Key Factors Underlying Ischemia-Reperfusion Injury in Human Liver Transplantation [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/comprehensive-and-combined-transcriptomic-and-proteomic-analysis-reveals-key-factors-underlying-ischemia-reperfusion-injury-in-human-liver-transplantation/. Accessed February 22, 2026.« Back to 2019 American Transplant Congress