Single Cell RNAseq Analysis of Human Renal Allograft Rejection Demonstrates Distinct Transcriptomic Profiles That Vary Based on the Type of Maintenance Immunosuppression
T. Shi1, A. Burg1, K. Roskin2, J. Rush3, B. Haraldsson3, A. Shields4, R. Alloway4, E. Woodle5, D. Hildeman1
1Immunobiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 2Biomedical Informatics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 3Novartis Pharmaceuticals AG, Basel, Switzerland, 4Internal Medicine, University of Cincinnati, Cincinnati, OH, 5Surgery, University of Cincinnati, Cincinnati, OH
Meeting: 2022 American Transplant Congress
Abstract number: 1285
Keywords: Biopsy, Genomics, Graft-infiltrating lymphocytes, Leukocytes
Topic: Basic Science » Basic Science » 12 - Immunosuppression & Tolerance: Preclinical & Translational Studies
Session Information
Session Name: Immunosuppression & Tolerance: Preclinical & Translational Studies
Session Type: Poster Abstract
Date: Monday, June 6, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
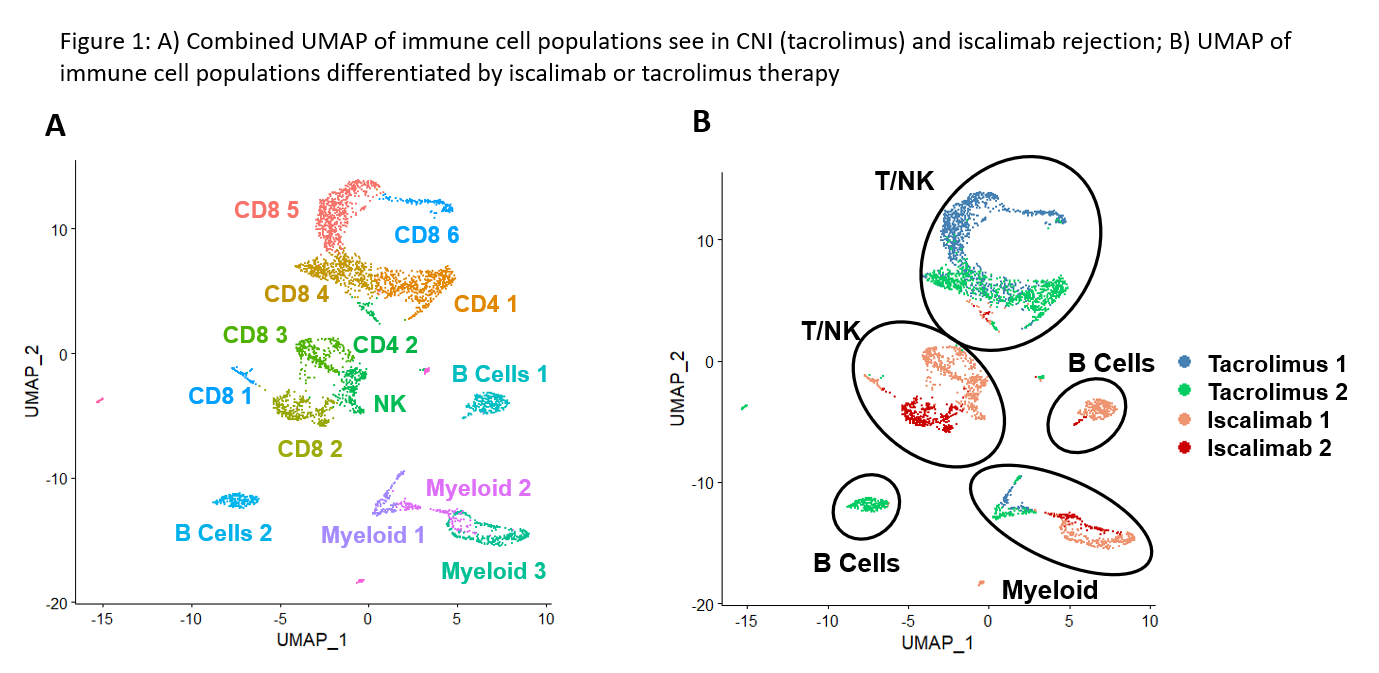
*Purpose: Rejection therapy has remained unchanged for 60 years and is associated with significant failure and infection rates. Development of new anti-rejection therapies requires mechanistic understanding of transplant rejection. Here, we employed single cell RNA sequencing (scRNAseq) to redefine renal allograft rejection (RAR) with single cell resolution.
*Methods: Biopsies and urine samples were obtained at the time of initial RAR from 4 patients on either iscalimab- (anti-CD40 antibody) or calcineurin inhibitor (CNI)-based maintenance immunosuppression (IS). Biopsies were digested at 4C with psychrophilic enzymes, and scRNAseq, including V(D)J sequencing, was performed on both biopsy and urine samples. Cell populations were analyzed based on their gene expression profiles using Seurat. Gene expression profiles were compared between infiltrating immune cells isolated from CNI vs iscalimab treated patients.
*Results: Analysis revealed similar frequencies of various immune cell populations within biopsies from patients rejecting under CNI vs iscalimab. Cells from iscalimab biopsies clustered similarly to each other, but distinctly from cells from CNI biopsies, indicating reproducible transcriptomic profiles for each IS therapy (Fig. 1). Myeloid cell populations between iscalimab- and CNI-treated patients cluster closely together and displayed similar gene expression profiles. Importantly, T cell and B cell populations under iscalimab clustered distinctly from those under CNI. Consistent with known effects of CD40 blockage, B cell analysis demonstrated naïve, transitional, and follicular profiles in CNI-treated patients, but not in iscalimab-treated patients. Ongoing work will further characterize differential gene expression in the T cell and myeloid cell populations between the CNI and iscalimab samples.
*Conclusions: Distinct gene expression profiles are observed in CD8+ T cells and B cells depending upon the type of IS therapy. These data suggest that the underlying biology of transplant rejection likely varies based on the nature of IS therapy, thereby emphasizing the importance for understanding rejection mechanisms under distinct IS regimens.
To cite this abstract in AMA style:
Shi T, Burg A, Roskin K, Rush J, Haraldsson B, Shields A, Alloway R, Woodle E, Hildeman D. Single Cell RNAseq Analysis of Human Renal Allograft Rejection Demonstrates Distinct Transcriptomic Profiles That Vary Based on the Type of Maintenance Immunosuppression [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/single-cell-rnaseq-analysis-of-human-renal-allograft-rejection-demonstrates-distinct-transcriptomic-profiles-that-vary-based-on-the-type-of-maintenance-immunosuppression/. Accessed February 18, 2026.« Back to 2022 American Transplant Congress