Gene Profiling to Distinguish Mechanisms of Liver Allograft Rejection and Acceptance in Cynomolgus Monkey
1Columbia Center for Translational Immunology, Columbia University, New York, NY, 2Department of Pediatrics, Columbia University, New York, NY, 3Departments of Pathology and Cell Biology, Columbia University, New York, NY, 4Department of Surgery, Columbia University, New York, NY
Meeting: 2022 American Transplant Congress
Abstract number: 1275
Keywords: Gene expression, Liver grafts, T cell graft infiltration, Tolerance
Topic: Basic Science » Basic Science » 12 - Immunosuppression & Tolerance: Preclinical & Translational Studies
Session Information
Session Name: Immunosuppression & Tolerance: Preclinical & Translational Studies
Session Type: Poster Abstract
Date: Monday, June 6, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
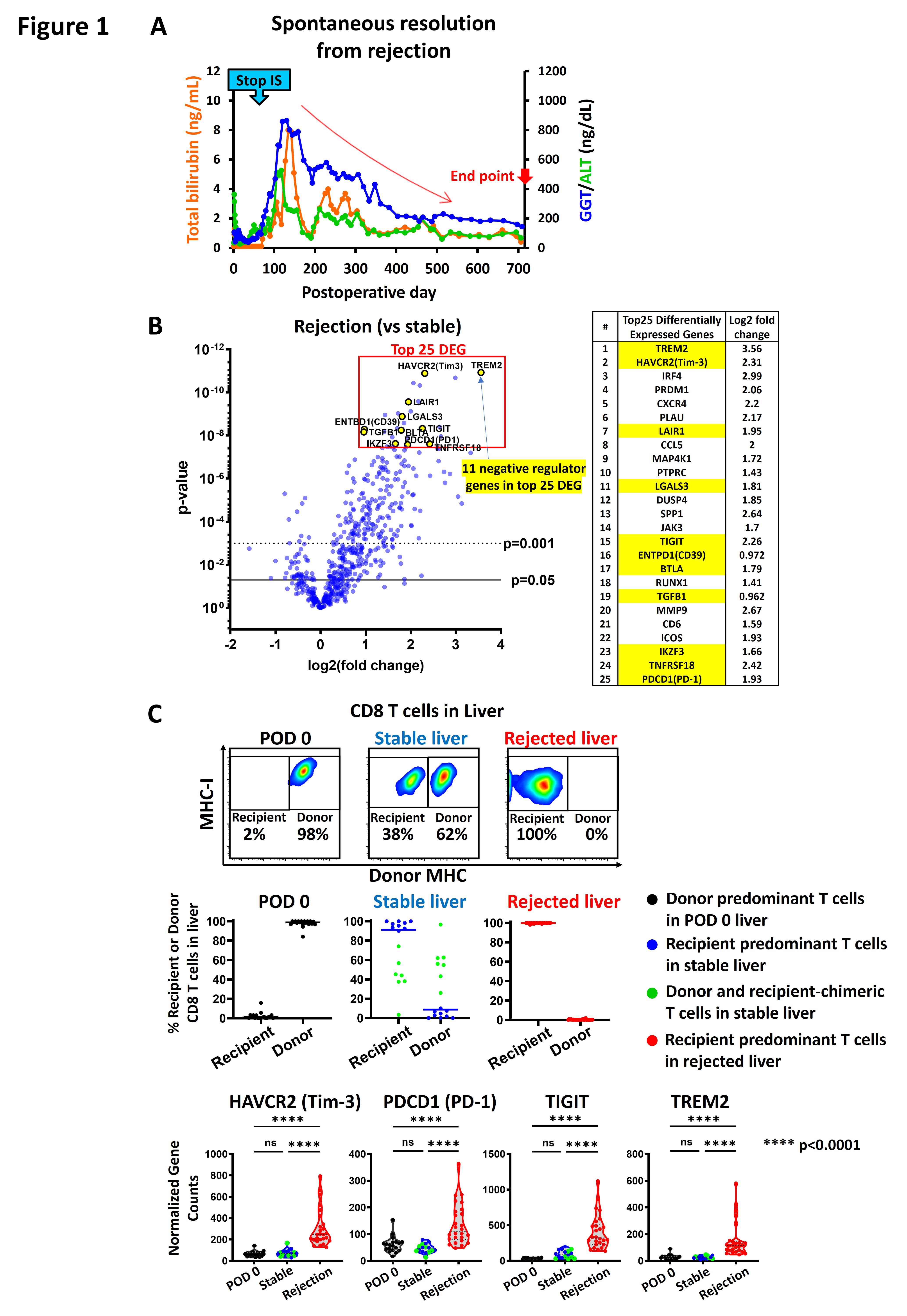
*Purpose: Liver is an immune regulatory organ. Spontaneous resolutions from rejection have been seen in our orthotopic cynomolgus monkey liver transplant models for tolerance induction in which all immunosuppression (IS) was stopped 2 months after transplantation (Figure1A). To elucidate the T cell inhibitory mechanism of the liver allograft in the monkey model, we performed gene profiling for 67 liver biopsies from 22 recipients.
*Methods: Subjects received 3 different protocols in addition to tacrolimus-based IS maintenance: 1) total body/thymic irradiation (TBI/TI) plus donor bone marrow transplantation (BMT); 2) TBI/TI without BMT; and 3) cyclophosphamide. Liver wedge biopsies were done on POD 0, in stable and rejection graft phases. 754 mRNA species in each liver sample were analyzed by nanoString.
*Results: 3 groups (POD 0, stable, and rejection) showed distinctive gene profiles. No difference in the gene profile was seen among IS protocols. Cytotoxic CD8 T cell score calculated with the gene set was higher in the rejected livers than POD 0 and stable livers, which was consistent with the % of CD8+ effector memory T cells in the liver analyzed by flow cytometry. Interestingly, negative regulator genes were highly ranked in differentially expressed genes (DEG) of the rejected livers. The top gene was TREM2, which was recently identified as a marker of immunosuppressive myeloid cells. Among the top 25 DEG, 10 were other negative regulators including T cell inhibitory receptors such as HAVCR2(Tim3), TIGIT, and PTCD1(PD-1), while they were not upregulated in the stable livers (Figure1B/C). Donor T cells were seen in some of the stable livers by flow cytometry. Surprisingly, no difference in expression of the genes encoding inhibitory receptors on T cells was seen between donor and recipient T cells in the stable liver (Figure1C). Not only individual genes but many negative regulation gene sets in the Gene Ontology Biological Process database were significantly enriched in the rejected livers.
*Conclusions: Liver allograft microenvironment may modulate T cell activation via inhibitory receptors regardless of self or donor T cells.
To cite this abstract in AMA style:
Sakai H, Berglund E, Chaudhry S, Kato Y, Duggan E, Weiner J, Alonso-Guallart P, Llore N, Brustle K, Stern J, Piegari B, Ekanayake-Alper D, Martinez M, Pierre G, Danton M, Kofman S, Ordanes D, Hajosi D, Lefkowitch J, Iuga A, Kato T, Griesemer A. Gene Profiling to Distinguish Mechanisms of Liver Allograft Rejection and Acceptance in Cynomolgus Monkey [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/gene-profiling-to-distinguish-mechanisms-of-liver-allograft-rejection-and-acceptance-in-cynomolgus-monkey/. Accessed February 10, 2026.« Back to 2022 American Transplant Congress