Renal Allograft Rejection: New Paradigms in the T Cell Landscape Revealed by Single Cell RNAseq Analysis
1Immunobiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 2Biomedical Informatics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 3Novartis Pharmaceuticals AG, Basel, Switzerland, 4Internal Medicine, University of Cincinnati, Cincinnati, OH, 5Surgery, University of Cincinnati, Cincinnati, OH
Meeting: 2022 American Transplant Congress
Abstract number: 372
Keywords: Genomics, T cell clones, T cell receptors (TcR), Tissue-specific
Topic: Basic Science » Basic Science » 02 - Acute Rejection
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 6:00pm-6:10pm
Presentation Time: 6:00pm-6:10pm
Location: Hynes Room 309
*Purpose: Defining the T cell landscape in renal allograft rejection (RAR) transcriptomically at the single cell level remains to be achieved. The purpose of the present study was to define distinct T cell populations involved in RAR.
*Methods: Renal allograft biopsy samples along with paired urine samples were collected from 9 patients during for cause biopsy procedures from kidney transplant patients. 5’ single cell RNA sequencing (scRNAseq) with paired TCRα/β sequencing (TCRseq) was performed on urine and biopsy samples that were digested at 4C using psychrophilic enzymes. Whole transcriptomic and TCR data was analyzed by Seurat and Loupe V(D)J browser, and individual T cell CDR3α/β clonotypes were identified. Expanded clones were defined as > 2 cells and 0.5% of T cell barcodes identified with identical CDR3α/β sequences.
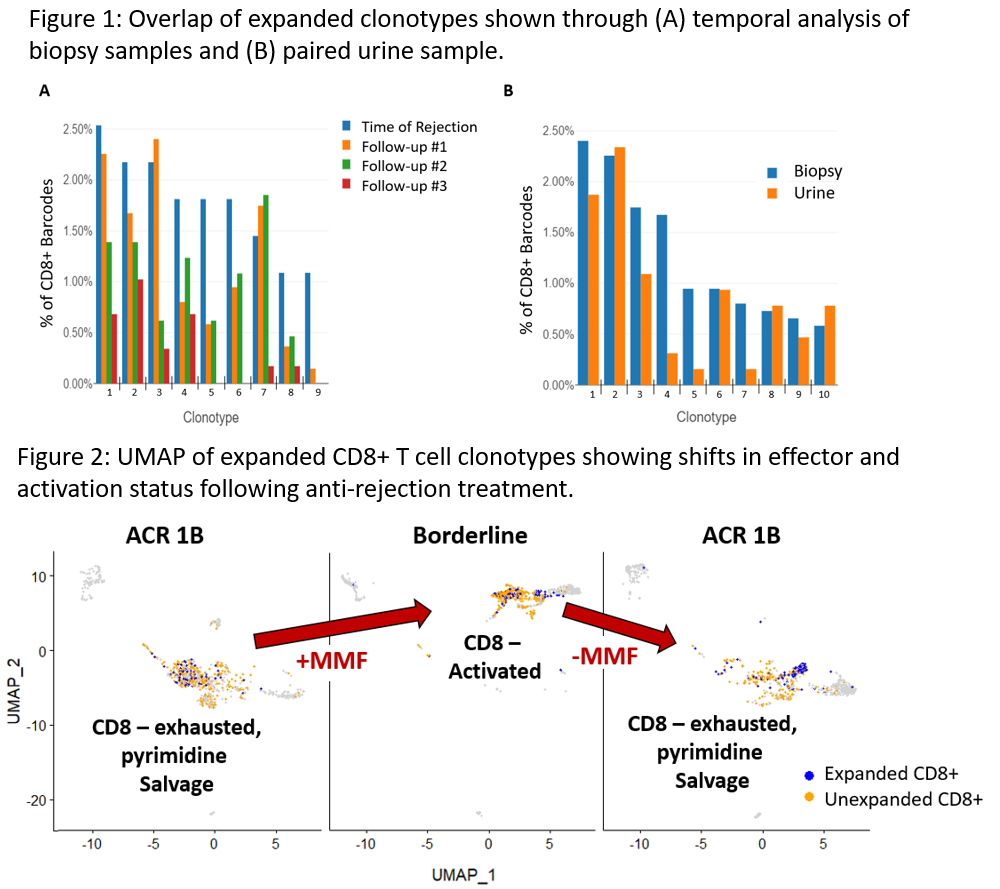
*Results: Samples obtained at the time of RAR diagnosis revealed an impressive degree of TCR restriction (avg. 14 expanded CD8+ clones/patient). Transcriptomic profiles of the expanded clones included markers associated with effector function, consistent with an ongoing alloresponse. To assess the possibility that the limited number of expanded clones was due to sampling bias, we analyzed 4 patients who had subsequent biopsies following RAR diagnosis. The subsequent biopsies demonstrated that >80% of the same clonally expanded T cells remained present (Fig 1a). Similarly, biopsy-paired urine samples demonstrated a substantial overlap in TCR clones with biopsy samples (Fig 1b). Interestingly, in patients with rejection reversal, clonal populations were largely eliminated and replaced with memory phenotype CD8+ T cells. In contrast, in patients with treatment resistant rejection, clonal populations were maintained. However, their gene expression profiles changed in response to anti-rejection therapies (Fig 2).
*Conclusions: 1) RAR is characterized by a remarkably limited number of distinct CD8+ T cell clones, 2) CD8+ T cell clones persist in ongoing rejection and disappear with rejection resolution, 3) CD8+ T cell clones isolated from simultaneous urine samples are highly similar to those isolated from biopsies, and 4) transcriptional profiles of clonally expanded cells shift in response to anti- rejection treatment.
To cite this abstract in AMA style:
Shi T, Burg A, Roskin K, Rush J, Haraldsson B, Shields A, Alloway R, Woodle E, Hildeman D. Renal Allograft Rejection: New Paradigms in the T Cell Landscape Revealed by Single Cell RNAseq Analysis [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/renal-allograft-rejection-new-paradigms-in-the-t-cell-landscape-revealed-by-single-cell-rnaseq-analysis/. Accessed February 16, 2026.« Back to 2022 American Transplant Congress