MiRNA as Regulator of Cell Death and Inflammatory Response in Hepatic Iri
1Surgery, University of Maryland, Baltimore, MD, 2Surgery, James D Eason Transplant Institute, Memphis, TN, 3Surgery, University of Maryland - Division of Surgical Science, Baltimore, MD
Meeting: 2021 American Transplant Congress
Abstract number: 595
Keywords: Genomics, Ischemia, Liver, Liver transplantation
Topic: Basic Science » Ischemia Reperfusion & Organ Rehabilitation
Session Information
Session Name: Ischemia Reperfusion & Organ Rehabilitation
Session Type: Poster Abstract
Session Date & Time: None. Available on demand.
Location: Virtual
*Purpose: Hepatic IRI represents a barrier to the use of available organs. Early graft dysfunction of donor livers is directly related to the severity of hepatic IRI. We aimed to identify upstream regulators of IRI pathways as targets for therapeutics interventions.
*Methods: High-throughput gene expression , miRNAs and DNA methylation were done in pre-implantation (L1) and post-reperfusion (L2) biopsies from DDLT recipients. After pre-processing and normalization, differential expressed genes (FDR 0.05, FC>2), miRNAs (FDR 0.05) and CpGs (FDR 0.05, delta>0.20) were identified between groups and at each time point. CpGs were mapped to genes. Integration of significant miRNAs and mRNAs was done using IPA.
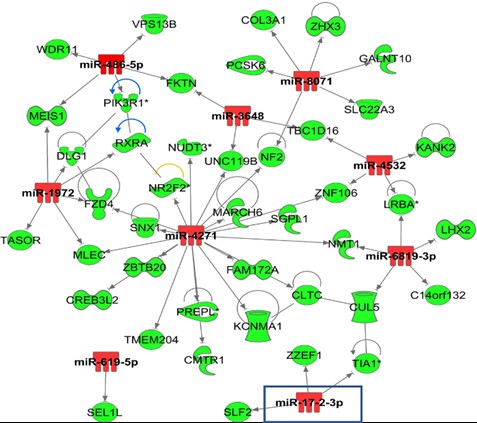
*Results: Patients presented different extent of IRI post-LT (high (n=20) and low (n=20) IRI groups). CIT was similar between groups (7.4 ± 1.8 vs. 7.1 ± 2.8, p=0.62). Steatosis (%) was statistically different between groups (% steatosis: 25 ± 11.8 vs. 4.1 ± 5.6 ± 3. 4, p<0.001). A total of 20 miRNAs in the high IRI group and 26 miRNAs in the low IRI group were statistically differentially expressed (FDR 0.05), of which 7 miRNAs were common to both groups. Differentially expressed miRNAs and mRNAs from same samples were integrated. Within the high IRI group 16 out of 20 differentially expressed miRNAs presented 70 gene targets also differentially expressed in the same samples and following the predicted directionality in expression (Fig. 1). For the low IRI low injury group, 15 out of 26 differentially expressed miRNAs presented 95 gene targets differentially expressed in the same tissue samples. The integrated list of DEGs from the high IRI group when enriched for functions showed activation of cell death (p= 2.9E-04, Z-score 4.7), growth failure (p= 1.4E-03; Z-score 3) and inhibition of transport of molecules (p= 3.23E-03; Z-score 3.02). Analysis from the low IRI injury group showed gene enriched functions mainly related to chemotaxis (p = 1.45E-12, Z-core 2.23), migration of neutrophils (p= 2.69E-05; Z-score 2.41), and T cell development (p= 6.33E-12; Z-score 2.42). Differentially expressed miRNAs were also presenting differentially methylated CpGs at their gene regulatory regions (i.e., MIR17 (TSS1500), MIRLET7A2 (TSS200), MIR144|MIR451 (TSS1500), C9orf3| MIR23B| MIR27B (3UTR|TSS1500).
*Conclusions: These preliminary integrative analyses support a role for miRNAs as regulators of pathways of donor organ damage in hepatic IRI and likely targets for intervention.
To cite this abstract in AMA style:
Bardhi E, Rousselle T, McDaniels J, Bontha S, Eason J, Maluf D, Mas V. MiRNA as Regulator of Cell Death and Inflammatory Response in Hepatic Iri [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/mirna-as-regulator-of-cell-death-and-inflammatory-response-in-hepatic-iri/. Accessed February 18, 2026.« Back to 2021 American Transplant Congress