A Single-Cell Transcriptome Atlas of 15 Adult Human Organs
1Organ Transplant Center, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China, 2The Cancer Centre of Sun Yat-Sen University, Guangzhou, China
Meeting: 2020 American Transplant Congress
Abstract number: D-345
Keywords: Gene expression, Lymphocytes, T cell receptors (TcR), Transcription factors
Session Information
Session Name: Poster Session D: Lymphocyte Biology: Signaling, Co-Stimulation, Regulation
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: The transcriptome diversity of cell types within and across tissues is of great importance for biological function of human beings. Here we conducted a systematic analysis of cell similarity and heterogeneity across multiple organs of healthy human adults
*Methods: We performed single-cell RNA, T cell receptor (TCR) and B cell receptor (BCR) sequencing on 15 organs from one adult male. The sequencing data was used to revealed annotation, similarities, differences and potential communications between different cell types
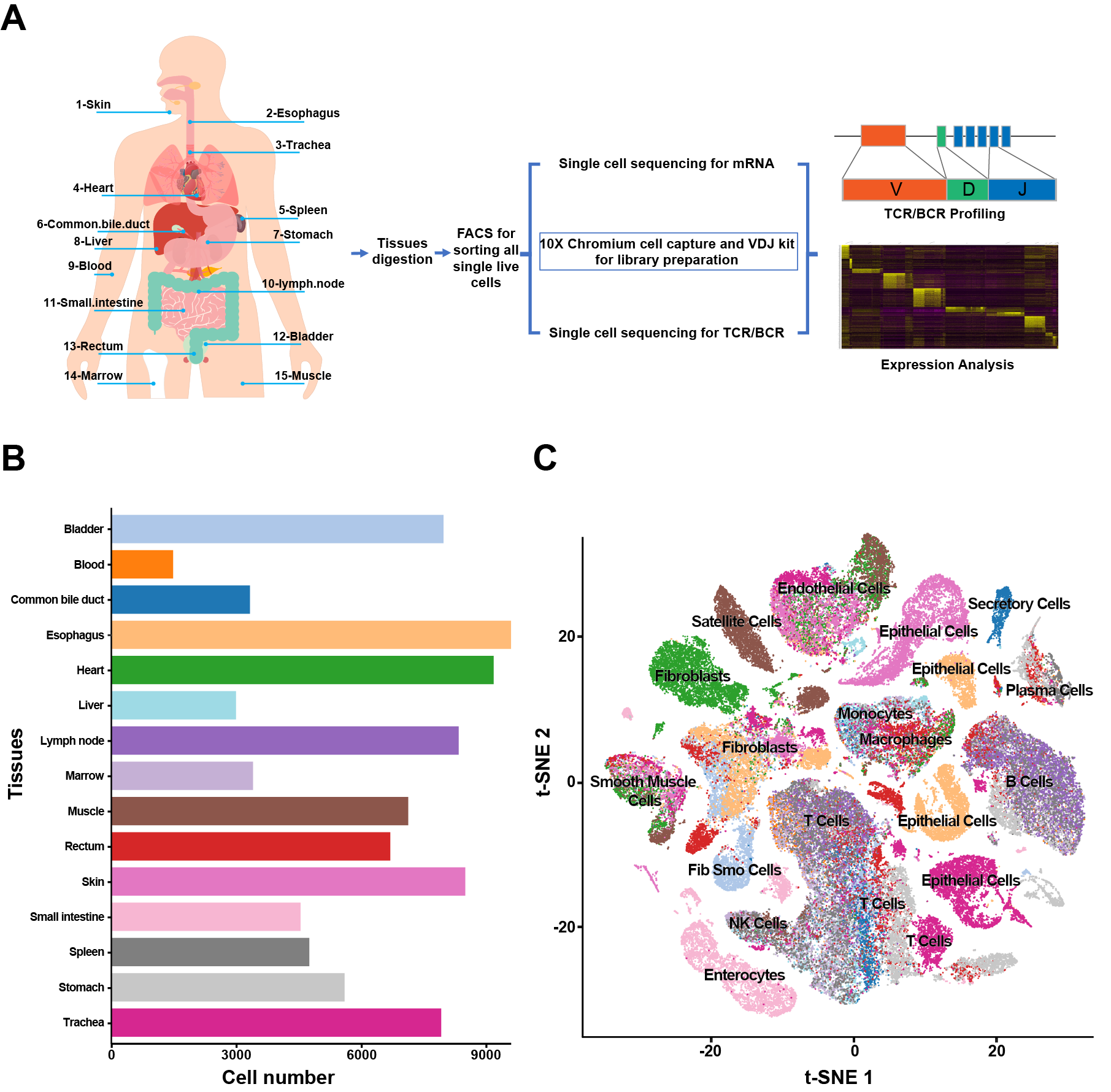
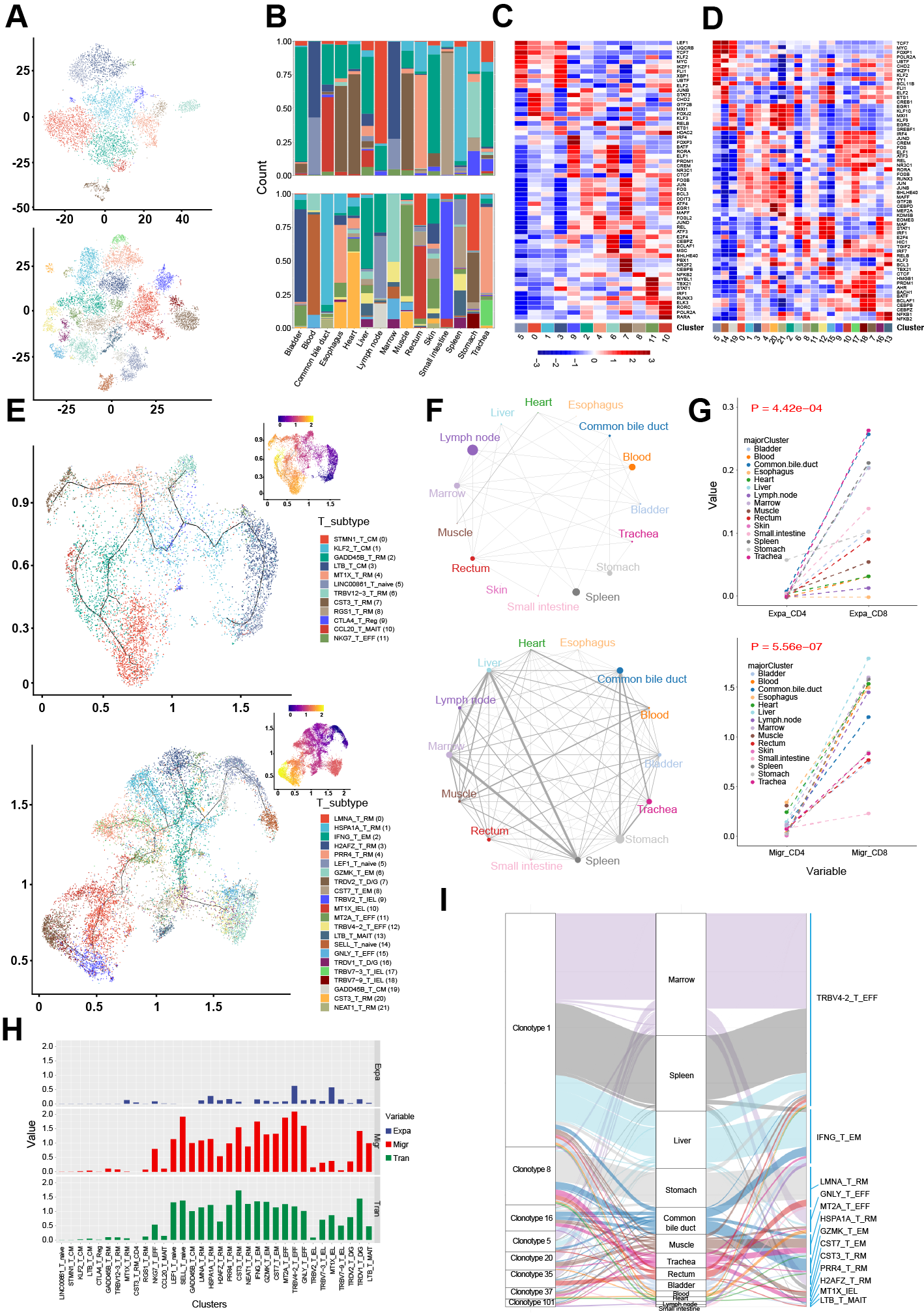
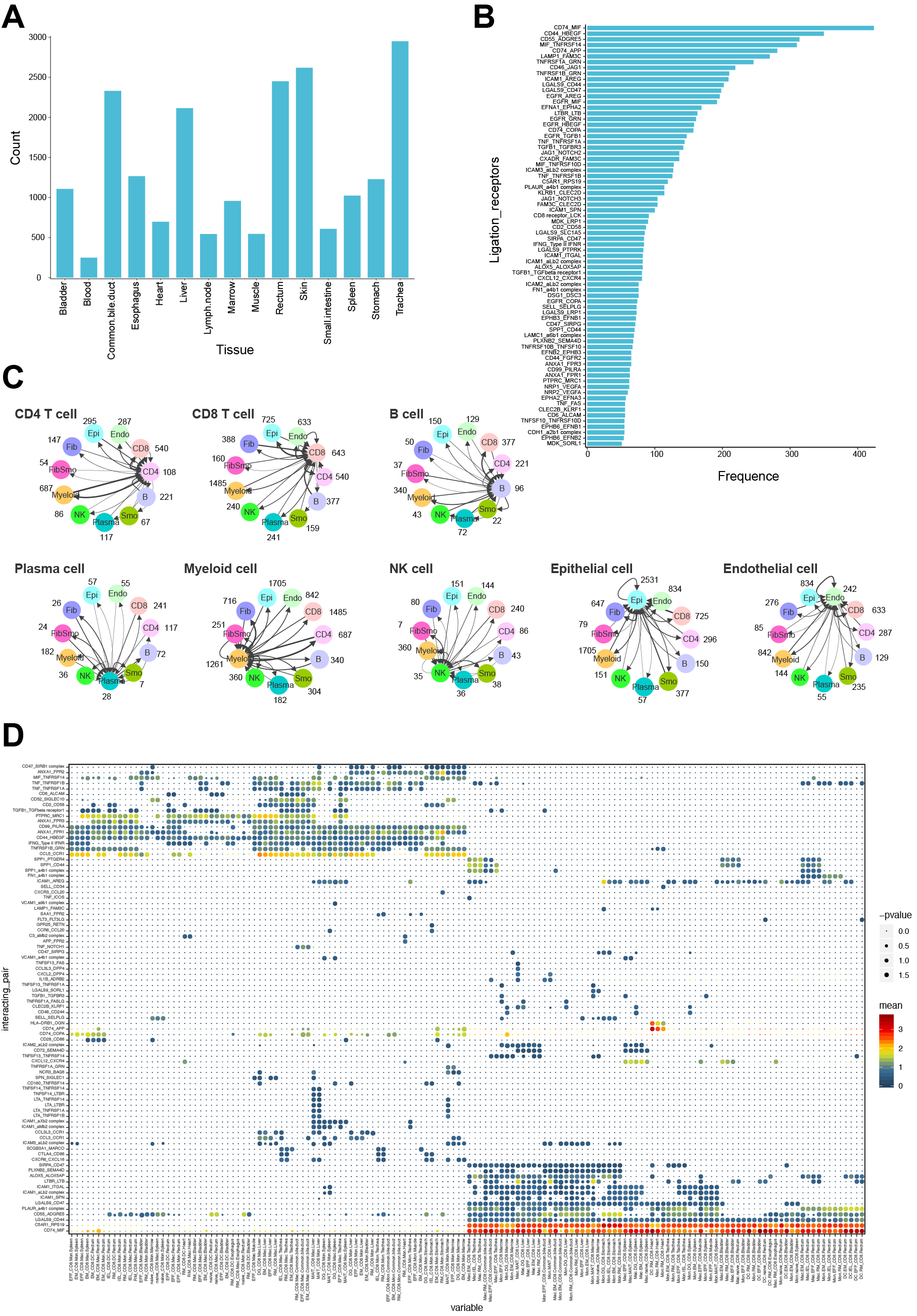
*Results: We isolated single cells using enzymatic digestion and selected all single live cells (Fig. 1A). 91393 cells were captured and sub-clustered into 43 subsets (Fig.1B & 1C). 20494 T cells were classified into CD4+ (11 clusters) and CD8+ T cells (22 clusters) according gene profiles (Fig. 2A & 2B). Trajectory and transcription factors analysis of CD4+ and CD8+ T cells revealed their special transcriptional regulation and differentiation mechanism. Meanwhile, TCR analysis of CD4+ T cell showed the different clone types and lower expansion ability than CD8+ (Fig. 2 C-I). We performed independent analyses on B cells, monocytes, macrophages, epithelial cells, endothelial cells, fibroblasts and smooth muscle cells. The molecular interactions and ligation-receptor pairs analysis revealed 468 different potential interactions in 15 tissues with 20694 combinations between cell types and established the broad intercellular communication networks within tissues (Fig. 3)
*Conclusions: Our dataset provides a resource for transcriptome characteristics of multiple organs at the single-cell level. This high-resolution adult human cellular atlas (AHCA) provides a foundation of knowledge to study biology of normal human cells and development of diseases
To cite this abstract in AMA style:
Guo Z, He S, Wang L, Liu Y, Xu J, Bei J, He X. A Single-Cell Transcriptome Atlas of 15 Adult Human Organs [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/a-single-cell-transcriptome-atlas-of-15-adult-human-organs/. Accessed February 17, 2026.« Back to 2020 American Transplant Congress