Assessment of Molecular Mechanisms of Non-Transplant Cardiac Hypertrophy in Cardiac Allograft Hypertrophy – An Exploratory Gene Array Study of Myocardial Biopsy Samples.
1Houston Methodist DeBakey Heart & Vascular Center, Houston, TX
2Houston Methodist Research Institute, Houston, TX.
Meeting: 2016 American Transplant Congress
Abstract number: B142
Keywords: Endomyocardial biopsy, Gene expression, Graft failure, Heart/lung transplantation
Session Information
Session Name: Poster Session B: Hearts and VADs in Depth - The Force Awakens
Session Type: Poster Session
Date: Sunday, June 12, 2016
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Halls C&D
Cardiac Allograft Hypertrophy (CAH) develops in up to 80% of patients as early as six months post-transplant, and is associated with decreased quality of life and decreased survival. We performed genomic analysis in 10 endomyocardial biopsies from 5 patients who underwent heart transplantation. Biopsies were taken at week 4 and week 20 post-transplant. We analyzed cardiac hypertrophy related genes within the epithelial to mesenchymal transition (EMT), collagen, angiotensin, fetal gene and IL6 dependent pathways. Gene expression at week 20 was correlated with LV mass measured by echocardiograpy, myocyte size, acute cellular rejection, C4D deposition, and interstitial fibrosis. The mean age was 59 ± 5 yrs (60% Caucasian, 60% male). 8/11 EMT pathway genes were downregulated in most patients. COL1A2, COL3A1, and COL5A1 genes were upregulated in most patients (3/5) with more than 2 fold increase, 8/12 angiotensin dependent hypertrophy pathway genes were upregulated in most patients. 4/7 NFAT/GATA4 family genes were upregulated in most patients (while 2 other genes had no change). Results were inconsistent for IL6 dependent hypertrophy pathway genes. Echo LV mass at week 20 had a significant correlation with the expression of SNAI1, (r=.92;p=0.026), Col3A1 (r=0.88,p=0.046), MAPK9 (r=0.89, p=0.038); myocyte size with MMP2 (r=0.82,p=0.046). While ACR had a significant correlation with only IL-6 (r=0.91,p=0.029) and NFAT C4 (r=0.92,p=0.026), C4D deposition had a significant correlation with MMP-9 (r=0.97, P=0.004), AGTRAP (r=0.92, p=0.023), IL-6R (0.096, p=0.008) and STAT 4 (r=0.98,p=0.003) genes. 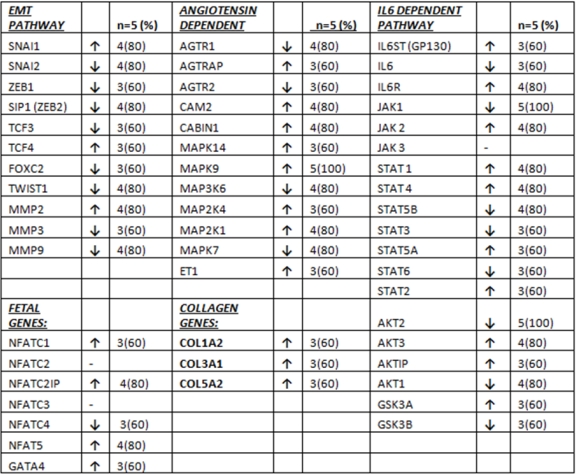
Our results show that early CAH could be caused by increased collagen gene expression and by activation of the angiotensin dependent hypertrophy pathway but not the IL6 dependent pathway. NFAT/GATA4 fetal genome pathways are also upregulated similar to non-transplanted cardiac hypertrophy mechanisms.
CITATION INFORMATION: Araujo-Gutiérrez R, Ibarra-Cortez S, Park M, Estep J, Ashrith G, Trachtenberg B, Torre-Amione G, Youker K, Bhimaraj A. Assessment of Molecular Mechanisms of Non-Transplant Cardiac Hypertrophy in Cardiac Allograft Hypertrophy – An Exploratory Gene Array Study of Myocardial Biopsy Samples. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Araujo-Gutiérrez R, Ibarra-Cortez S, Park M, Estep J, Ashrith G, Trachtenberg B, Torre-Amione G, Youker K, Bhimaraj A. Assessment of Molecular Mechanisms of Non-Transplant Cardiac Hypertrophy in Cardiac Allograft Hypertrophy – An Exploratory Gene Array Study of Myocardial Biopsy Samples. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/assessment-of-molecular-mechanisms-of-non-transplant-cardiac-hypertrophy-in-cardiac-allograft-hypertrophy-an-exploratory-gene-array-study-of-myocardial-biopsy-samples/. Accessed March 5, 2026.« Back to 2016 American Transplant Congress
