Whole-Exome Sequencing Identifies PMS1 and DYRK1A Genes Contributing to Occurrence of Liver Cancer
Tongji hospital, Huazhong university, Organ Transplantation, Wuhan, China
Meeting: 2019 American Transplant Congress
Abstract number: A142
Keywords: Genomics, Hepatocellular carcinoma, Liver transplantation
Session Information
Session Name: Poster Session A: Biomarkers, Immune Monitoring and Outcomes
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: Liver cancer is one of the most frequent cancers in China and the accurate landscape of liver cancer, especially of HBV-infected liver cancer in China need to be explained.
*Methods: We selected a total of 26 adult patients between 2016 and 2017 who were diagnosed with liver cancer and subsequently received allogeneic liver transplantation. We collected tumor tissues and case-matched peripheral blood for whole-exome sequencing (WES). We profiled the somatic mutations spectrum of the samples. Furthermore, we used CHASM software to predict the new driver genes in Chinese population. In addition, we found a germline mutation in one patient with hepatocellular carcinoma using whole-exome sequencing.
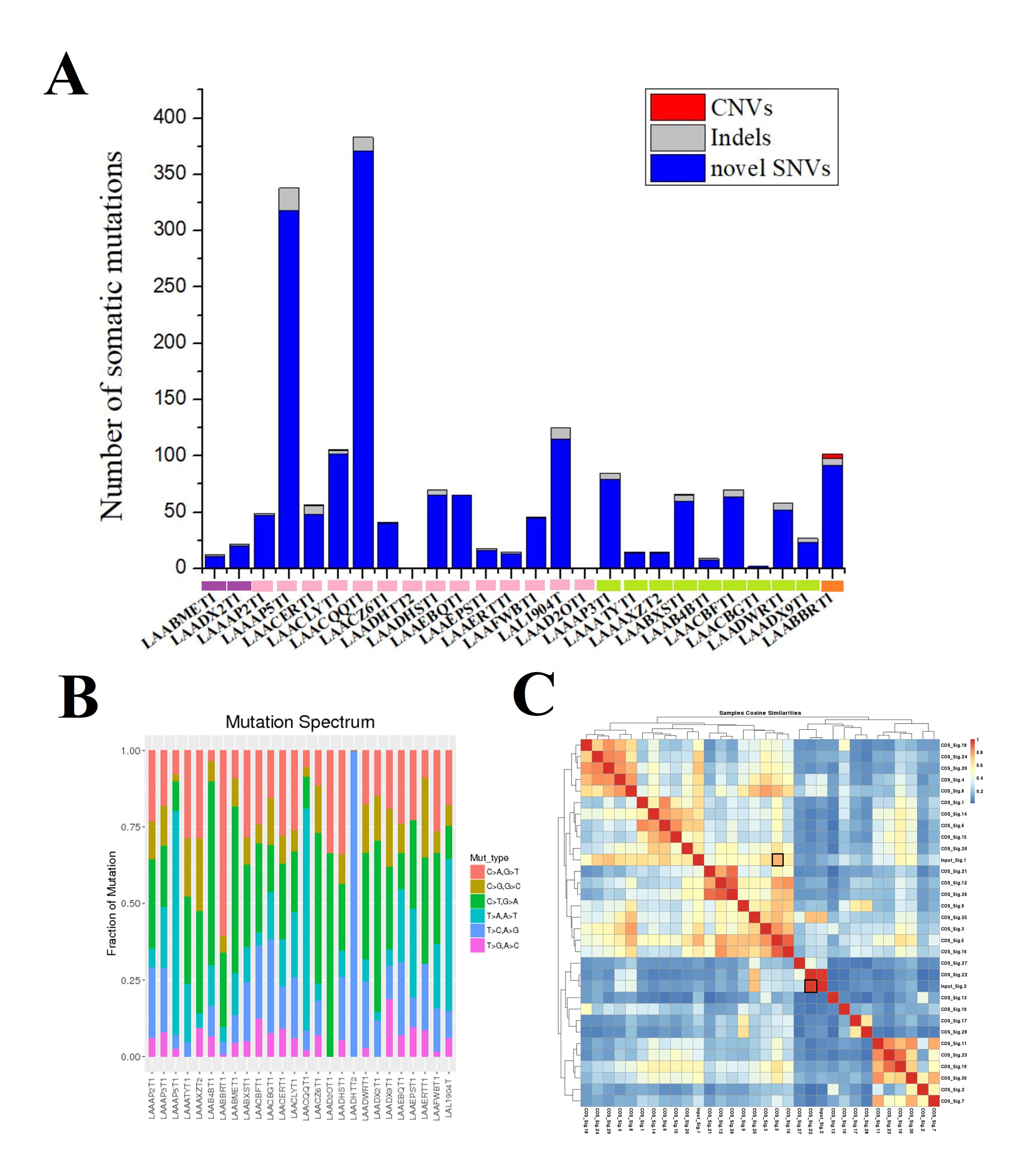
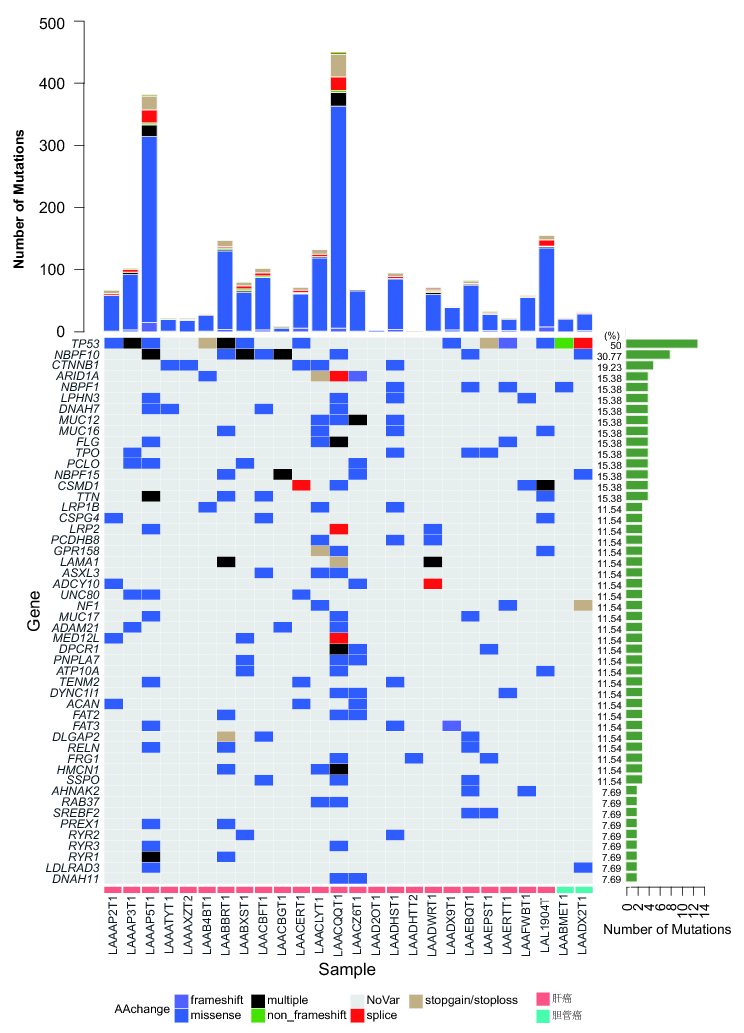
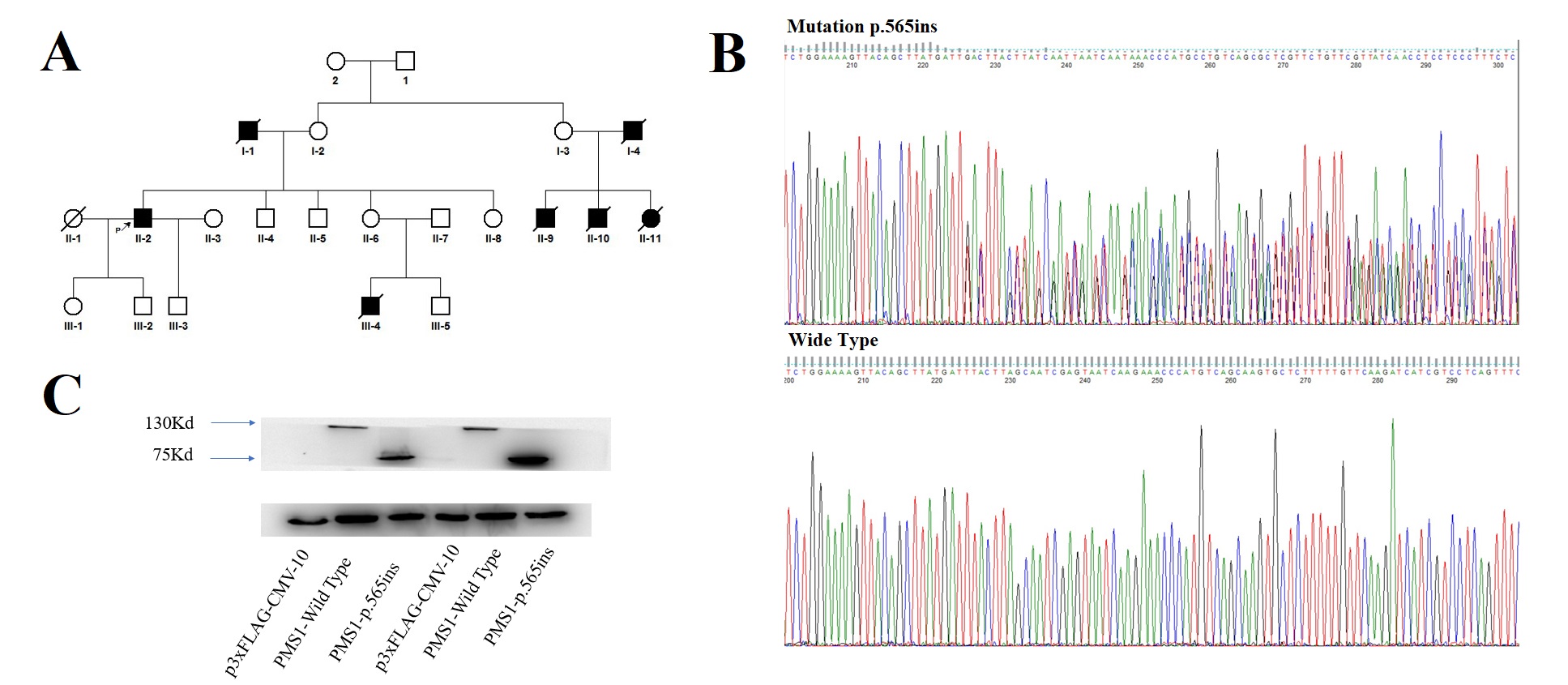
*Results: Overall, we identified 1796 mutations including novel SNVs, indels and CNVs were identified (Fig. 1A). Mutation spectrum analysis of CNVs showed that the frequency of mutation type T>G (A>C) and C>G (G>C) were relatively low (Fig. 1B). We compared the mutation signatures of our results with Catalogue of Somatic Mutations in Cancer (COSMIC) Database and the results showed that the mutation signatures in our cohort got high similarity compared with standard COS_Sig.22 (Cosine similarity score >0.90, Fig. 1C). Next, there were 41 genes with 3 or more nonsynonymous somatic mutations (Fig 2). We observed that mutations in gene TP53 was the most frequently occurred (50%) in our cohort. In addition, the frequency of mutations in gene NBPF10 and CTNNB1 were also high (30.77% and 19.23%, respectively). Furthermore, we confirmed 5 novel genes PRDM2, CTNNB1, TSC2, DYRK1A and NR4A3 with FDR<0.1 after adjustment of Benjamini-Hochberg (Table 1). Among them, gene DYRK1A was considered as a new driver gene in liver cancer. In addition, we found a novel mutation p.Leu565X in gene PMS1. Mutations of PMS1 has never been reported in liver cancer. The genogram was showed in Fig 3A. Among 20 members in the family, 7 members were occurred osteocarcinoma, colon cancer, gastric carcinoma or liver cancer. We verified the mutation p.Leu565X of gene PMS1 by Sanger sequencing (Fig 3B). The Western Blot analysis result showed that the mutation cause the cut-off of protein (Fig 3C). Based on the results, we considered that PMS1 was a novel pathogenic gene of hereditary hepatocellular carcinoma.
*Conclusions: The somatic mutations spectrum in liver cancer in China is characteristic and novel genes are associated with liver cancer in Chinese population.
| Gene Name | Number of variants | CHASM gene score | CHASM gene FDR | TARGET |
| PRDM2 | 1 | 0.934 | 0.05 | NA |
| CTNNB1 | 5 | 0.944 | 0.05 | WNT inhibitors |
| TSC2 | 2 | 0.802 | 0.1 | Everolimus, Temsirolimus, MTOR inhibitors |
| DYRK1A | 2 | 0.910 | 0.1 | NA |
| NR4A3 | 1 | 0.892 | 0.1 | NA |
To cite this abstract in AMA style:
Zhao Y, Yang B, Chen D, Wei L, Chen Z. Whole-Exome Sequencing Identifies PMS1 and DYRK1A Genes Contributing to Occurrence of Liver Cancer [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/whole-exome-sequencing-identifies-pms1-and-dyrk1a-genes-contributing-to-occurrence-of-liver-cancer/. Accessed February 27, 2026.« Back to 2019 American Transplant Congress