Spatially Resolved Molecular Profiling of Endothelial Cells in Antibody-Mediated Kidney Transplant Rejection
UAB School of Medicine, Birmingham, AL
Meeting: 2022 American Transplant Congress
Abstract number: 478
Keywords: Biopsy, Endothelial cells, Gene expression, Rejection
Topic: Basic Science » Basic Science » 16 - Biomarkers: -omics and Systems Biology
Session Information
Session Name: Biomarkers: -omics and Systems Biology
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 7, 2022
Session Time: 3:30pm-5:00pm
 Presentation Time: 4:30pm-4:40pm
Presentation Time: 4:30pm-4:40pm
Location: Hynes Room 304 / 306
*Purpose: Endothelial cells are key targets of alloimmune injury in antibody-mediated rejection (ABMR), but are challenging to study in situ using high-throughput methods. We aimed to spatially resolve the whole transcriptome in CD31+ kidney transplant endothelial cells affected by ABMR.
*Methods: We used the NanoString GeoMx Digital Spatial Profiling (DSP) platform to generate spatially resolved whole transcriptome data from CD31+ kidney transplant endothelial cells in situ. We used formalin-fixed, paraffin-embedded slides taken from two archived kidney transplant biopsies (ABMR and Stable) that were residual to clinical use. We labeled CD31+ endothelial cells in glomerular, peritubular capillary, and arteriolar compartments in each biopsy using immunofluorescence. We then used the segmentation function of DSP to release barcode sequences corresponding to the whole transcriptome from multiple areas of CD31+ endothelial cells in situ. We quantified gene expression using next-generation sequencing then mapped the data back to the original locations in situ. We compared endothelial gene expression patterns between ABMR and Stable biopsies. We input differentially expressed genes into DAVID to identify biological pathways involved in endothelial cells affected by ABMR in situ.
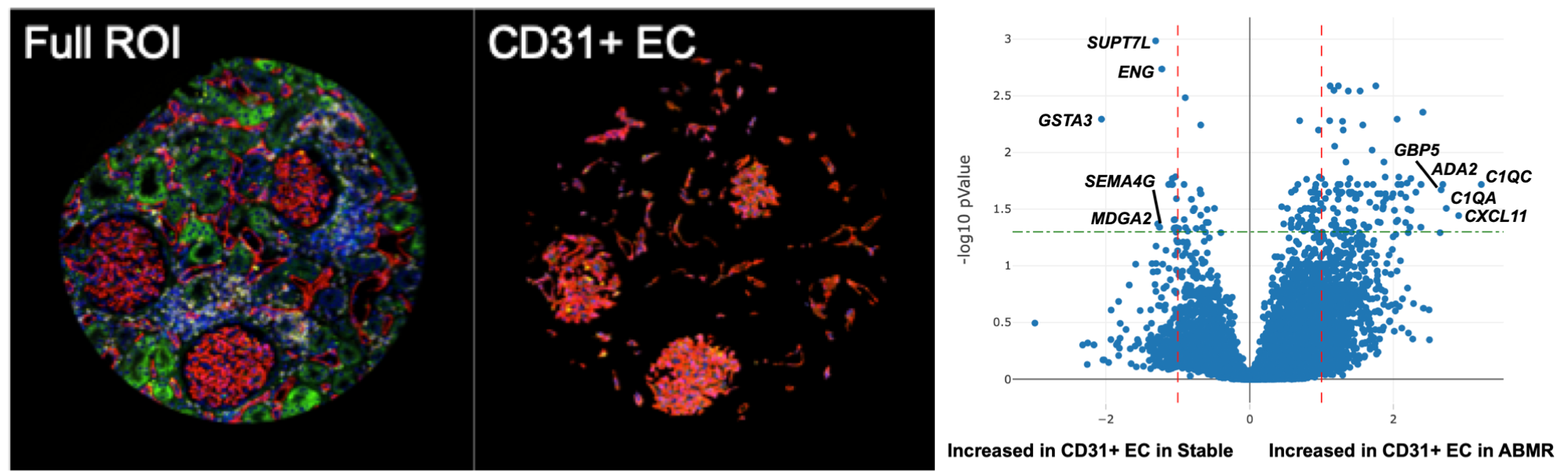
*Results: We profiled 18 areas of endothelium from an ABMR biopsy and 7 areas of endothelium from a Stable biopsy (Figure). We found 96 genes with increased expression and 17 genes with decreased expression in ABMR vs. Stable endothelium (Figure), which revealed enrichment for complement activation and innate immune pathways in ABMR and enrichment of angiogenesis and VEGF signaling pathways in Stable. Of the 96 genes in ABMR-affected endothelium, only CXCL11 was previously identified in multiple ABMR classifiers in the Banff 2017 prime gene list.
Figure. (Left) Geometric profiling by DSP to select an area of the biopsy enriched with microvasculature. (Middle) Segmentation with DSP isolates CD31+ endothelium from the same area. (Right) Volcano plot shows differential gene expression between ABMR and Stable endothelium; top differentially expressed genes are noted in black.
*Conclusions: Spatially resolved molecular profiling using NanoString’s DSP platform identified several novel significant genes upregulated in CD31+ kidney transplant endothelial cells affected by ABMR. We speculate that spatially resolved molecular profiles of endothelial injury in situ can be used to build diagnostic classifiers for ABMR that outperform existing classifiers based on bulk molecular profiling.
To cite this abstract in AMA style:
Melendez-Ferro M, Kelly DR, Seifert ME. Spatially Resolved Molecular Profiling of Endothelial Cells in Antibody-Mediated Kidney Transplant Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/spatially-resolved-molecular-profiling-of-endothelial-cells-in-antibody-mediated-kidney-transplant-rejection/. Accessed March 9, 2026.« Back to 2022 American Transplant Congress