Single Cell RNA Sequencing Identifies Primarily Terminal Effector and Exhausted CD8+T Cells in Chronically Rejected Human Kidney Allografts
1Department of Surgery, Houston Methodist Hospital, Houston, TX, 2School of Biomedical Informatics, The University of Texas Health Science Center at Houston, Houston, TX
Meeting: 2022 American Transplant Congress
Abstract number: 115
Keywords: Gene expression, Kidney transplantation, Rejection, T cells
Topic: Clinical Science » Kidney » 47 - Kidney Complications: Immune Mediated Late Graft Failure
Session Information
Session Name: Kidney Complications: Chronic Antibody Mediated Rejection & Immune Mediated Late Graft Failure
Session Type: Rapid Fire Oral Abstract
Date: Sunday, June 5, 2022
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:50pm-6:00pm
Presentation Time: 5:50pm-6:00pm
Location: Hynes Room 302
*Purpose: Chronic renal allograft rejection often leads to irreversible damage, despite intensification of immunosuppression (ISN). T cell phenotypes in this clinical setting are not well described. Evaluating T cell function on a transcriptional level, through single cell RNA-seq, reveals unique T cell subsets, such as stemness (naïve, effector precursor), terminal effector, or exhaustion, in kidney transplant recipients with chronic rejection.
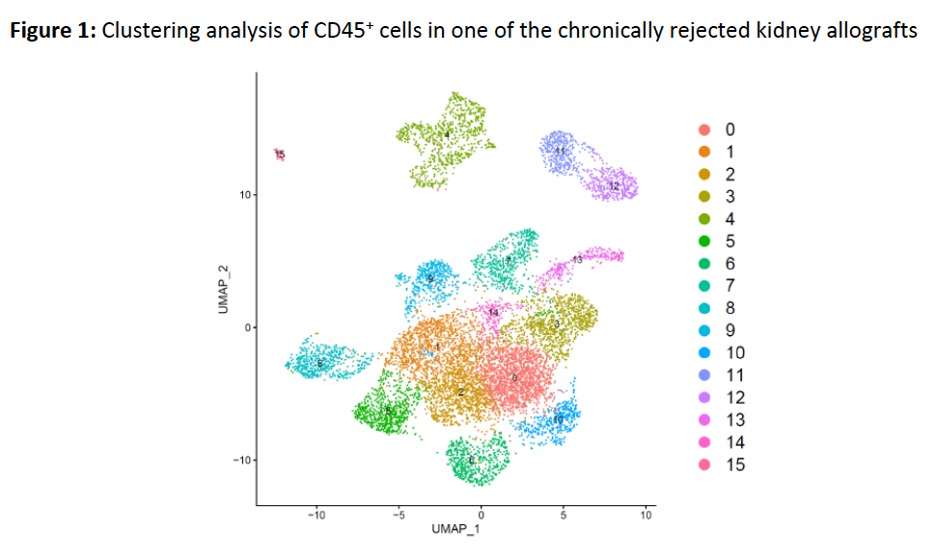
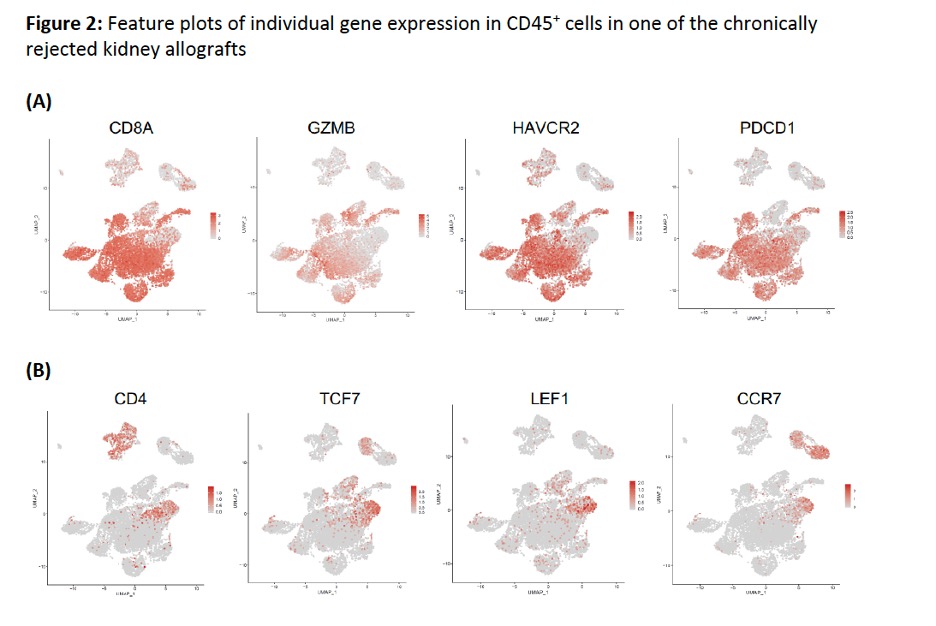
*Methods: Peripheral blood mononuclear cells (PBMC) and allograft biopsies from 3 recipients off ISN showing chronic cellular/antibody mediated rejection were analyzed and compared. Living CD45+ cells were isolated, followed by 10x Genomics scRNA-seq and bioinformatic analysis using R platform. Cell clustering by uniform manifold approximation and projection (UMAP) visualization (Figure 1), and feature plot visualization of individual gene expression of transcription factors, cytokines/effector molecules, and cell surface receptors were used (Figure 2).
*Results: The majority of CD4+ and CD8+ T cells in PBMC highly expressed T cell stemness associated genes (TCF7, LEF1, CCR7, SELL, CD127, BCL2, etc; data not shown). However, most CD8+ T cells infiltrated in chronically rejected allografts appeared to lose stemness, becoming TCF7–GZMBhi effector T cells or TCF7–PDCD1hiHAVCR2hi exhausted T cells (Figure 2A). Only some allograft CD4+ T cells maintained TCF7hi stemness (Figure 2B).
*Conclusions: Through scRNA-seq, we identified a phenotypic transition from stemness to terminal effector or exhaustion among CD8+ T cells in chronic allograft rejection. Similarly, CD4+ T cells appeared to lose stemness, suggesting a transition to an effector state. Further investigation into the regulatory processes of this dynamic cellular change is necessary to identify therapeutic targets that can prevent irreversible allograft damage in chronic kidney rejection.
To cite this abstract in AMA style:
Yi SG, Zou D, Dai Y, Zhao Z, Chen W, Gaber A. Single Cell RNA Sequencing Identifies Primarily Terminal Effector and Exhausted CD8+T Cells in Chronically Rejected Human Kidney Allografts [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/single-cell-rna-sequencing-identifies-primarily-terminal-effector-and-exhausted-cd8t-cells-in-chronically-rejected-human-kidney-allografts/. Accessed July 13, 2025.« Back to 2022 American Transplant Congress