Predictors of Graft Survival at the Time of a Kidney Transplant Indication Biopsy
1Hannover Medical School, Hannover, Germany
2Alberta Transplant Applied Genomics Centre, Edmonton, Canada
3Laboratory Medicine and Pathology, University of Alberta, Edmonton, Canada
4Medicine, University of Alberta, Edmonton, Canada.
Meeting: 2018 American Transplant Congress
Abstract number: 322
Keywords: Biopsy, Gene expression, Graft survival
Session Information
Session Name: Concurrent Session: Kidney Complications: Late Graft Failure
Session Type: Concurrent Session
Date: Monday, June 4, 2018
Session Time: 4:30pm-6:00pm
 Presentation Time: 5:42pm-5:54pm
Presentation Time: 5:42pm-5:54pm
Location: Room 6E
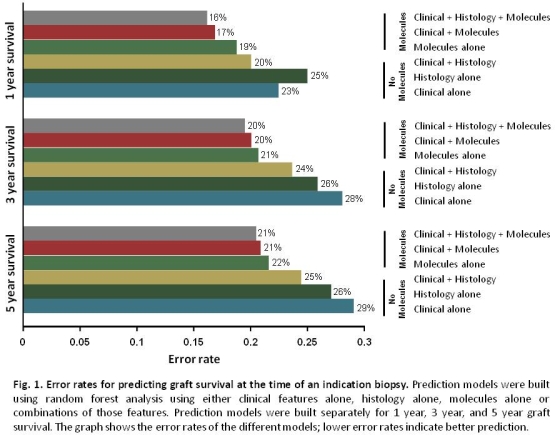
We predicted kidney transplant graft survival after indication biopsy in 884 patients. Analysis was performed using random forests, a multivariate machine learning method to assess if data from the biopsy (histology/molecules) adds predictive value to clinical data.
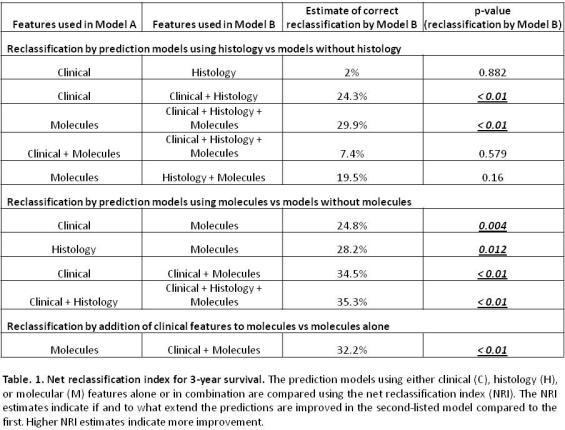
Survival predictions using either histology or clinical information were similar but were improved by combining both. Molecules were always better predictors than clinical or histology data alone or combined (Fig.1&2).
Using clinical variables alone, eGFR, time of biopsy post-transplant, and proteinuria were the most important predictors; DSA status was not important in multivariate analysis. When clinical and histology variables were combined, the best predictors were clinical features (eGFR, proteinuria), followed by fibrosis (ci) and glomerular double contours (cg). Inflammatory lesions (i, t, ptc, g), DSA, and histologic diagnosis were not important multivariate predictors. In models including molecules, the best independent predictors were molecular scores for injury (ci, AKI), or late-stage ABMR (cg, late-stage ABMR score) and molecules representing proteinuria or eGFR, while clinical and histologic features were less important.
Thus biopsies add predictive accuracy to clinical data, and significant improvement is gained by using molecular data compared to histology. Survival predictions can be made with molecular data alone, and are improved only slightly by the addition of clinical and histologic data. Many variables that are significant in univariate analysis (e.g. DSA, diagnosis, rejection lesions i, t, ptc, g) drop out in multivariate analyses when more relevant molecular predictors related to injury and ABMR stage are included.
CITATION INFORMATION: Einecke G., Reeve J., Halloran P. Predictors of Graft Survival at the Time of a Kidney Transplant Indication Biopsy Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Einecke G, Reeve J, Halloran P. Predictors of Graft Survival at the Time of a Kidney Transplant Indication Biopsy [abstract]. https://atcmeetingabstracts.com/abstract/predictors-of-graft-survival-at-the-time-of-a-kidney-transplant-indication-biopsy/. Accessed March 10, 2026.« Back to 2018 American Transplant Congress