Molecular Analysis of Rejection and Injury in Human Liver Transplant Biopsies: First Results of the INTERLIVER Study
University of Alberta, Edmonton AB, AB, Canada
Meeting: 2019 American Transplant Congress
Abstract number: 46
Keywords: Biopsy, Liver, Liver transplantation, Rejection
Session Information
Session Name: Concurrent Session: Liver: Immunosuppression and Rejection
Session Type: Concurrent Session
Date: Sunday, June 2, 2019
Session Time: 2:30pm-4:00pm
 Presentation Time: 3:42pm-3:54pm
Presentation Time: 3:42pm-3:54pm
Location: Room 312
*Purpose: The relationship of histologic liver transplant biopsy assessments to actual rejection and injury is unclear. Recent progress in molecular reclassification of kidney, heart, and lung transplant rejection suggests that microarray phenotyping of liver biopsies would be useful and potentially transformative.
*Methods: We prospectively studied 102 liver transplant biopsies (90% for indications) from USA, Canada, Europe, and Australia with gene expression microarrays (INTERLIVER ClinicalTrials.gov NCT03193151). Following strategies successful in other organs, we used 453 rejection-associated transcripts (RATs; derived in kidneys, validated in hearts and lungs) in unsupervised archetypal (AA) and principal component analyses (PCA).
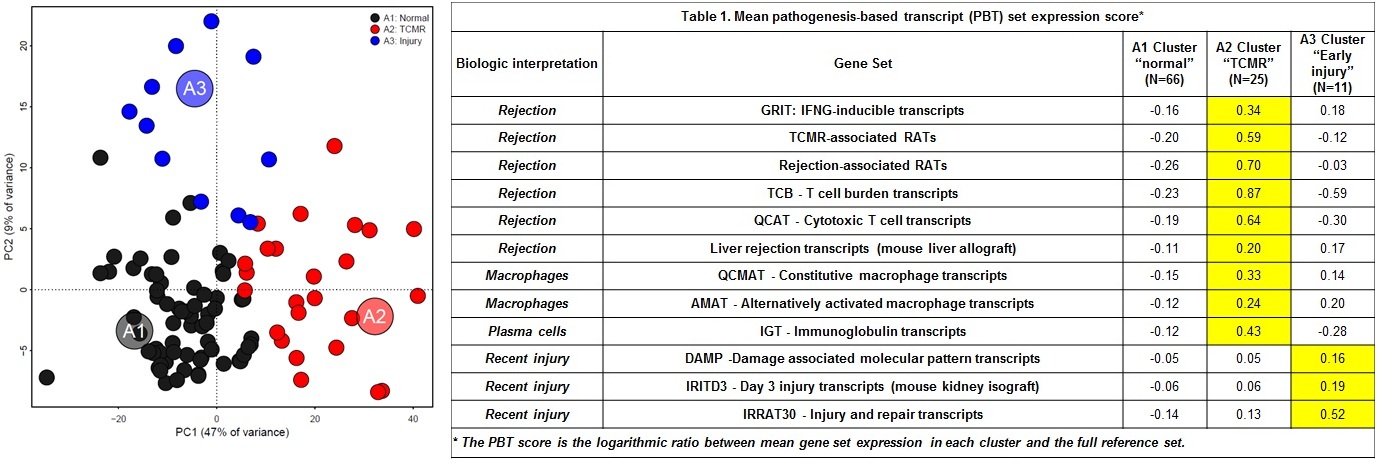
*Results: Every biopsy yielded high quality RNA for microarray analysis. In PCA based on RAT expression, Principal component 1 (PC1) explained 47% of variance, PC2 9%. AA identified 3 archetypes (A1normal, A2TCMR, and A3injury) and scored every biopsy for similarity to each archetype: S1normal, S2TCMR, and S3injury (Figure 1). S1normal anti-correlated and PC1 correlated with IFNG effects. S2TCMR and PC1 correlated with T cell-mediated rejection (TCMR) transcripts. PC1 and S2 correlated with histologic portal triaditis, compatible with TCMR. S3injury correlated with transcripts induced in injury models (e.g. SERPINB8), and anti-correlated with time post-transplant (ρS=-0.63) i.e. reflecting donation-implantation injury. Biopsies clustered as 66 normal, 25 TCMR, or 11 early injury by their highest archetype score. Median days from transplant to biopsy were A1 normal=1148, A2 TCMR=295, A3 injury=4. We studied the relationship of scores to annotated gene sets reflecting mechanisms (Table 1). Rejection and injury gene sets were low in A1normal. A2TCMR biopsies had highest effector T cell, macrophage, and IFNG-inducible transcripts, similar to TCMR in other organs. A3injury was high in transcripts that reflect injury and cellular damage (e.g. DAMPs). No biopsies had molecular changes similar to ABMR in other organs.
*Conclusions: MMDx-liver reveals at least three groups characterized by their relationships to previously annotated genes: normal, TCMR, and early injury. The incidence of biopsies with molecular TCMR-like changes (25%) was higher than previously reported in kidney and heart indication biopsies (<10%), which has interesting implications. No ABMR phenotype emerged but ABMR was not clinically suspected in these biopsies. Further studies will be required to characterize liver ABMR and other conditions e.g. steatosis.
To cite this abstract in AMA style:
. Molecular Analysis of Rejection and Injury in Human Liver Transplant Biopsies: First Results of the INTERLIVER Study [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/molecular-analysis-of-rejection-and-injury-in-human-liver-transplant-biopsies-first-results-of-the-interliver-study/. Accessed February 9, 2026.« Back to 2019 American Transplant Congress