Identification of Novel Gene Targets Inducing Resistance in Liver Ischemia Reperfusion Injury by CRISPR Screening
Dept of Surgery, Transplant Research Institute, James D. Eason Transplant Institute, University of Tennessee Health Science Center, Memphis, TN
Meeting: 2020 American Transplant Congress
Abstract number: 194
Keywords: Genomics, Hepatocytes, Ischemia, Screening
Session Information
Session Name: Basic: Ischemia Reperfusion & Organ Rehabilitation I
Session Type: Oral Abstract Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:45pm
 Presentation Time: 3:39pm-3:51pm
Presentation Time: 3:39pm-3:51pm
Location: Virtual
*Purpose: Liver transplantation (LT) is the best treatment option for patients with end-stage liver disease (ESLD). However, the gap between available donors and recipients in need continues increasing. Consequently, transplant centers are using high-risk donors that are known to be associated with exacerbated ischemia reperfusion injury (IRI )and worse outcomes. However, the molecular mechanisms associated with liver IRI need further study. This study aimed to identify novel target genes in IRI that can be manipulated by available drugs or gene editing.
*Methods: In vitro liver IRI was modeled by using two different strategies: 1) chemical induction with antimycin A and 2) hypoxia chamber for hepatic cell lines, HepG2. We used the same cell line in genome-wide CRISPR screening. We used Brunello pooled CRISPR library which can target more than 72,000 gRNA as a forward genetic screening tool in our established in vitro IRI model. Cas9 expressing HepG2 cells were transduced with gRNA virus at MOI:0.3 to assure single gene knock out in each individual cell. Cells that survived after IRI were collected by flow cytometry and used for genomic DNA isolation. DNAs from control and treated cells were used for next generation sequencing (NGS) to evaluate the frequency distribution of each gRNA. We used “MAGeCK” integrative analysis pipeline to identify target genes.
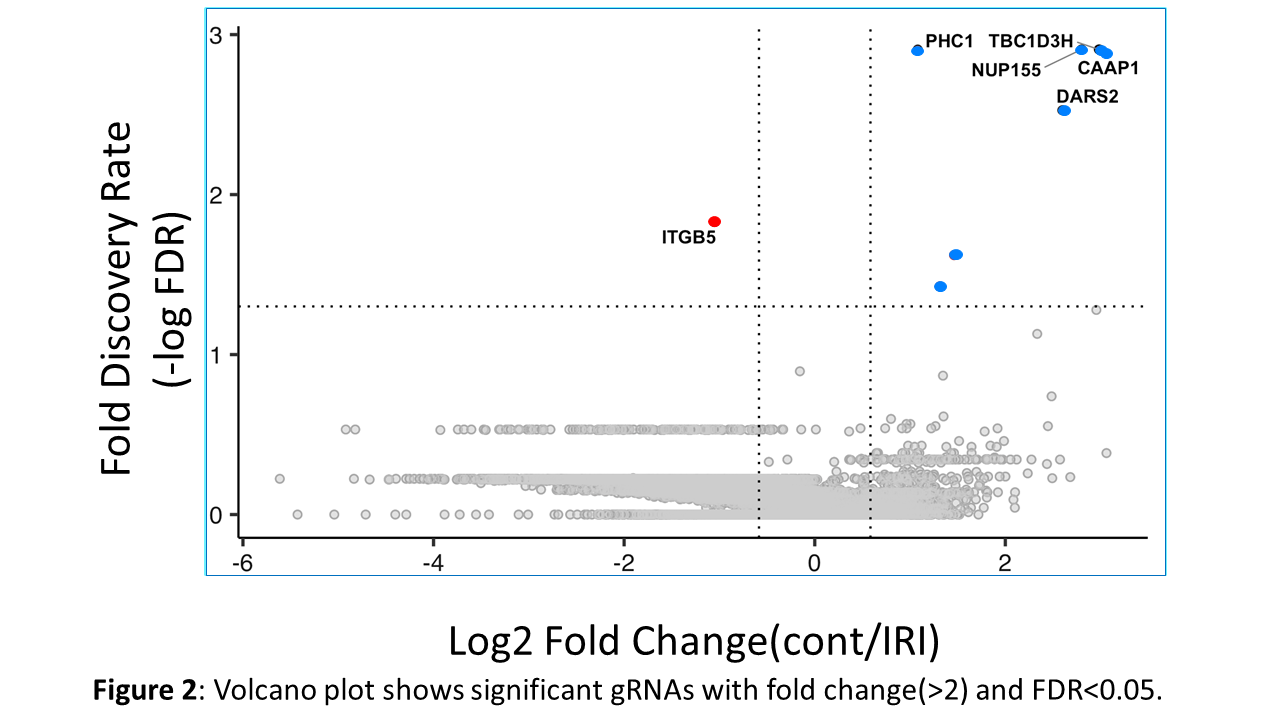
*Results: Our initial results demonstrate that chemical hypoxia model was more effective for cell death once cells were exposed to the oxygen and nutrient rich media, Fig 1. We identified 8 significant genes ; one out of 8 showed enriched gRNA profile and 7 out of 8 displayed depleted gRNAs profile after the IRI (p < 10*e-5 and FDR < 0.05, Fig 2). One of the depleted gene called “caspase activity and apoptosis inhibitor 1(CAAP1)” has potential to induce liver cell resistance (less cell death) when exposed to IRI.
*Conclusions: This is the first study that demonstrates the utility and applicability of CRISPR based library screening in an in vitro IRI model to identify novel target genes.
To cite this abstract in AMA style:
Kuscu C, Wolen AR, Kuscu C, Watkins C, Bajwa A, Eason JD, Maluf D, Mas V. Identification of Novel Gene Targets Inducing Resistance in Liver Ischemia Reperfusion Injury by CRISPR Screening [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/identification-of-novel-gene-targets-inducing-resistance-in-liver-ischemia-reperfusion-injury-by-crispr-screening/. Accessed February 23, 2026.« Back to 2020 American Transplant Congress