Genome Wide Non-HLA Alloimmunity Contributes to Graft Loss after Kidney Transplantation
Division of Nephrology and Dialysis, Medical University of Vienna, Vienna, Austria
Department of Nephrology, Institute for Clinical and Experimental Medicine, Prague, Czech Republic
Transplant Institute, University of Pennsylvania, Philadelphia, PA.
Meeting: 2018 American Transplant Congress
Abstract number: 508
Keywords: Allorecognition, Graft failure, Immunogenicity, Kidney transplantation
Session Information
Session Name: Concurrent Session: Kidney Complications: Antibody & Late Outcomes
Session Type: Concurrent Session
Date: Tuesday, June 5, 2018
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:30pm-4:42pm
Presentation Time: 4:30pm-4:42pm
Location: Room 4B
Background
Chronic rejection is the main cause of renal transplant loss after the first year and has mostly been attributed to alloimmune responses against human leukocyte antigens (HLA). Epidemiological data, however, suggests a contribution of non-HLA related alloimmune responses. Candidate antigen approaches relying on predefined minor histocompatibility antigens did not provide convincing evidence for the importance of non-HLA related alloimmune responses. The contribution of non-HLA donor recipient mismatches in protein coding genes on a genome wide basis has not yet been assessed.
Methods
We analyzed 479 kidney transplant recipients and the respective deceased donors from two transplant centers in Vienna and Prague with a median follow up of 6.5 years. Genotyping was performed using the Affymetrix Axiom Tx v1 GWAS Array (Affymetrix, Santa Clara, CA, USA) as part of the iGeneTrain consortium. Raw genotypes were phased with SHAPEIT and imputed with IMPUTE2 using GoNL and 1KG data as reference panels. The Ensembl Variant Effect Predictor was used for SNP annotation. We used a Cox PH model to analyze the association of non-synonymous single nucleotide mismatches with graft survival.
Results
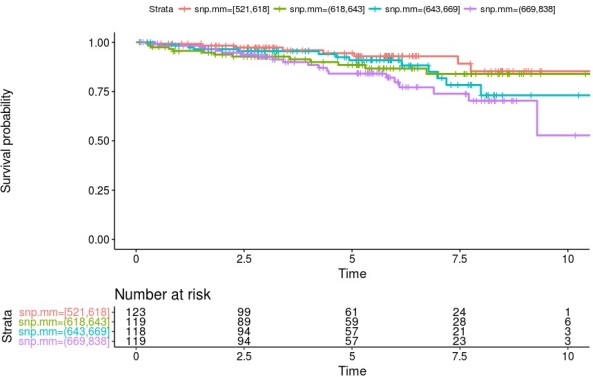
The analysis showed that the load of nsSNP MM in membrane and secreted proteins is associated with a statistically significant elevation in risk for death censored graft loss. This elevated risk (hazard ratio 10% per 10 nsSNP MM) remains after adjustment for HLA incompatibility *HLA-A, -B, -DR). Stratification of patients according to their number of nsSNPs into quartiles showed that especially kidney transplant recipients with a high number of mismatches have a lower graft survival probability (figure 1).
Conclusion
Genome wide genetic incompatibility in protein coding proteins (non-HLA) contributes to graft loss after kidney transplantation independently of HLA mismatches.
CITATION INFORMATION: Reindl-Schwaighofer R., Heinzel A., Kainz A., Jelencsics K., Viklicky O., Böhmig G., Fischer G., Keating B., Oberbauer R. Genome Wide Non-HLA Alloimmunity Contributes to Graft Loss after Kidney Transplantation Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Reindl-Schwaighofer R, Heinzel A, Kainz A, Jelencsics K, Viklicky O, Böhmig G, Fischer G, Keating B, Oberbauer R. Genome Wide Non-HLA Alloimmunity Contributes to Graft Loss after Kidney Transplantation [abstract]. https://atcmeetingabstracts.com/abstract/genome-wide-non-hla-alloimmunity-contributes-to-graft-loss-after-kidney-transplantation/. Accessed March 6, 2026.« Back to 2018 American Transplant Congress