Extended HLA Typing of Kidney Transplant Pairs is Necessary to Assign Presence of Donor-Specific Anti-HLA Antibodies
1Microbiology, Immunology and Transplantation, KU Leuven, Leuven, Belgium, 2HILA, Rode Kruis Vlaanderen, Leuven, Belgium
Meeting: 2020 American Transplant Congress
Abstract number: A-285
Keywords: Allorecognition, Epitopes, HLA antibodies, Major histocompatibility complex (MHC)
Session Information
Session Name: Poster Session A: Histocompatibility and Immunogenetics
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: The need for extended high-resolution (HR) HLA genotyping in solid organ transplantation is currently widely debated due to higher costs and questioned clinical importance. Here we evaluated the impact of different HLA genotyping levels on the assignment of the presence of DSA. Additionally, we investigated whether imputation of HR genotypes based on low-resolution (LR) genotypes could be used to correctly assign the presence of DSA.
*Methods: We included 1000 single kidney transplant pairs transplanted in one center between 2004 and 2014, initially typed at LR split antigen level. The presence of anti-HLA antibodies was assessed by Luminex. The whole cohort was retyped at HR level for 11 HLA loci using NGS. HR HLA imputation was performed using HaploStats.
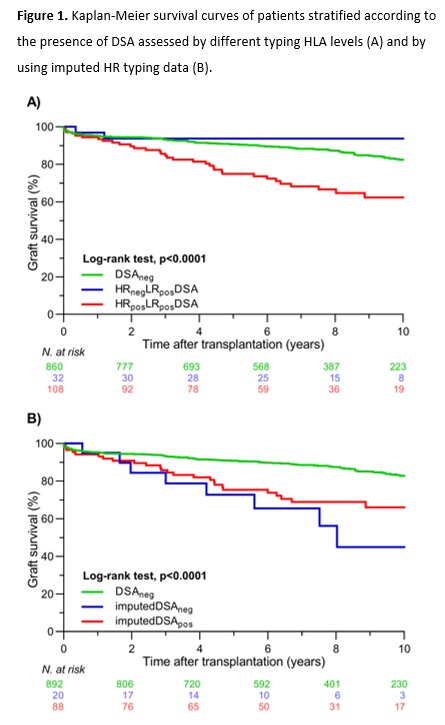
*Results: We detected pretransplant anti-HLA antibodies in 262/1000 recipients. Based on LR HLA typing donor data, serology or SSP for HLA-A/B/C/DRB1/DQB1, and Sanger sequencing for DPB1, pretransplant DSA were identified in 140 patients and absent in 860 patients (DSAneg group). With extended HLA typing data of both donors and patients, we confirmed the presence of DSA in 108/140 (77.1%) patients (HRposLRposDSA group) and excluded DSA in 32/140 patients (22.9%) (HRnegLRposDSA group). Kaplan-Meier curves showed that 10-year graft survival rates were similar between the DSAneg and HRnegLRposDSA groups (82.4% vs. 93.8%; p=0.27), which were both significantly better than graft survival in the HRposLRposDSA group (62.3%; p<0.001) (Figure1A). The main reasons for DSA misclassification were lack of 2nd field donor typing 23/40 (57.5%), incomplete typing of the donor 10/40 (25.0%), and incomplete typing of the patient 7/40 (17.5%). Analyzed per HLA molecule, the majority of misclassified DSAs were against DQ 18/40 (45.0%). Finally, when we used imputed HR HLA typing results, we found that in 19/23 (82.6%) of the misclassified DSA that required 2nd field HLA typing results of HRnegLRposDSA group DSA could be correctly assigned by imputation. However, when we analyzed the presence of DSA in HRposLRposDSA group using imputed results, in 20/108 (18.5%) patients, DSA would be wrongly excluded (imputedDSAneg) (Figure1B).
*Conclusions: Our study illustrates that extended HR HLA typing of the donor-recipient pairs is needed for correct assessment of DSA. Although imputation of HR genotypes from LR typing improves the assessment of DSA, significant misclassification occurs, and warrants caution in using imputed HLA results for clinical and research purposes.
To cite this abstract in AMA style:
Senev A, Sandt VVan, Naesens M, Emonds M. Extended HLA Typing of Kidney Transplant Pairs is Necessary to Assign Presence of Donor-Specific Anti-HLA Antibodies [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/extended-hla-typing-of-kidney-transplant-pairs-is-necessary-to-assign-presence-of-donor-specific-anti-hla-antibodies/. Accessed March 4, 2026.« Back to 2020 American Transplant Congress