Discovery and Validation of Blood Genomic Signatures of Acute Rejection in Liver Transplant Recipients
1Div of Gastroenterology & Comprehensive Tx Cnt, Chicago, IL, 2NIAID, Bethesda, MD, 3Rho, Chapel Hill, NC, 4UPMC, Pittsburgh, PA, 5Baylor, Dallas, TX, 6Mount Sinai, New York, NY, 7Mayo Arizona, Phoenix, AZ, 8Case Western, Cleveland, OH, 9Cleveland Clinic, Cleveland, OH, 10Comprehensive Transplant Center, Chicago, IL, 11Scripps, San Diego, CA
Meeting: 2019 American Transplant Congress
Abstract number: D135
Keywords: Gene expression, Genomic markers, Liver grafts
Session Information
Session Name: Poster Session D: Liver: Immunosuppression and Rejection
Session Type: Poster Session
Date: Tuesday, June 4, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
*Purpose: Biomarker profiles diagnostic of acute rejection (AR) could enhance the management of liver transplant (LT) recipients. Our aim was to discover and validate blood genomic signatures specifically diagnostic and predictive of AR in LT recipients.
*Methods: For discovery, LT recipients at our center (NU) had blood collected for mRNA expression at the time of liver biopsy and characterized as AR, ADNR (acute dysfunction/no rejection) and TX (no biopsy but non-viral recipient with normal liver tests). For validation, recipients enrolled in the multi-center NIAID CTOT-14 prospective study had serial blood collections prior to and at the time of AR, ADNR, and TX, as well as after AR treatment. Using random forest methods, the training NU set generated a locked classification model for AR vs. TX that was then validated on the CTOT-14 cohort.
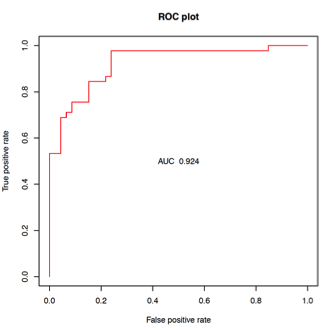
*Results: 46 AR, 38 ADNR, and 45 TX NU blood samples were analyzed for discovery. High predictive accuracy was obtained in differentiating AR from TX (AUC 0.92; Figure, Table) at diagnosis. In the CTOT-14 validation cohort, this locked model containing 36 gene probes and optimal probability threshold from the discovery cohort had reasonable AUC, accuracy, specificity and negative predictive value for AR (N=14) vs. TX (N=50) (Table). The sensitivity and positive predictive value were lower, supporting the test’s main value in ‘ruling out’ AR. When ADNR samples were analyzed on the same model/threshold, the majority (n=23; 82%) classified as TX and the minority as AR (n=5, 16%). We are currently analyzing samples prior to AR that might be useful to predict AR vs. other phenotypes, as well as after AR to monitor response to AR therapy. These results will be available at the time of presentation.
*Conclusions: We have discovered and validated blood diagnostic genomic signatures unique to AR in LT recipients. Current analyses are ongoing to determine the clinical utility of these biomarkers in predicting AR and in monitoring after treatment, allowing for serial monitoring that may improve immunosuppression optimization.
| AUC | Accuracy | Sensitivity | Specificity | PPV | NPV | |
| NU Discovery | 0.92 | 0.80 | 0.76 | 0.84 | 0.58* | 0.93* |
| CTOT14 Validation | 0.72 | 0.81 | 0.50 | 0.90 | 0.58 | 0.87 |
| *Prevalence-adjusted |
To cite this abstract in AMA style:
Levitsky J, Bridges N, Odim J, Brown M, Ikle D, Armstrong B, Demetris A, Asrani S, Klintmalm G, Schiano T, Moss A, Chavin K, Miller C, Kandpal M, Zhao L, Kurian S, Abecassis M. Discovery and Validation of Blood Genomic Signatures of Acute Rejection in Liver Transplant Recipients [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/discovery-and-validation-of-blood-genomic-signatures-of-acute-rejection-in-liver-transplant-recipients/. Accessed June 30, 2025.« Back to 2019 American Transplant Congress