Development of an M2 Macrophage Signature in Spontaneously Accepted Kidney Allografts
1Center for Transplantation Sciences, Massachusetts General Hospital, Boston, MA, 2Department of Surgery, Boston Children's Hospital, Boston, MA, 3Department of Pathology, Center for Transplantation Sciences, Massachusetts General Hospital, Boston, MA, 4Department of Surgery, Center for Transplantation Sciences, Massachusetts General Hospital, Boston, MA
Meeting: 2022 American Transplant Congress
Abstract number: 1238
Keywords: Gene expression, Genomics, Kidney transplantation, Tolerance
Topic: Basic Science » Basic Science » 08 - Innate Immunity; Chemokines, Cytokines, Complement
Session Information
Session Name: Innate Immunity; Chemokines, Cytokines, Complement
Session Type: Poster Abstract
Date: Monday, June 6, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:30pm-7:00pm
Presentation Time: 5:30pm-7:00pm
Location: Hynes Hall C
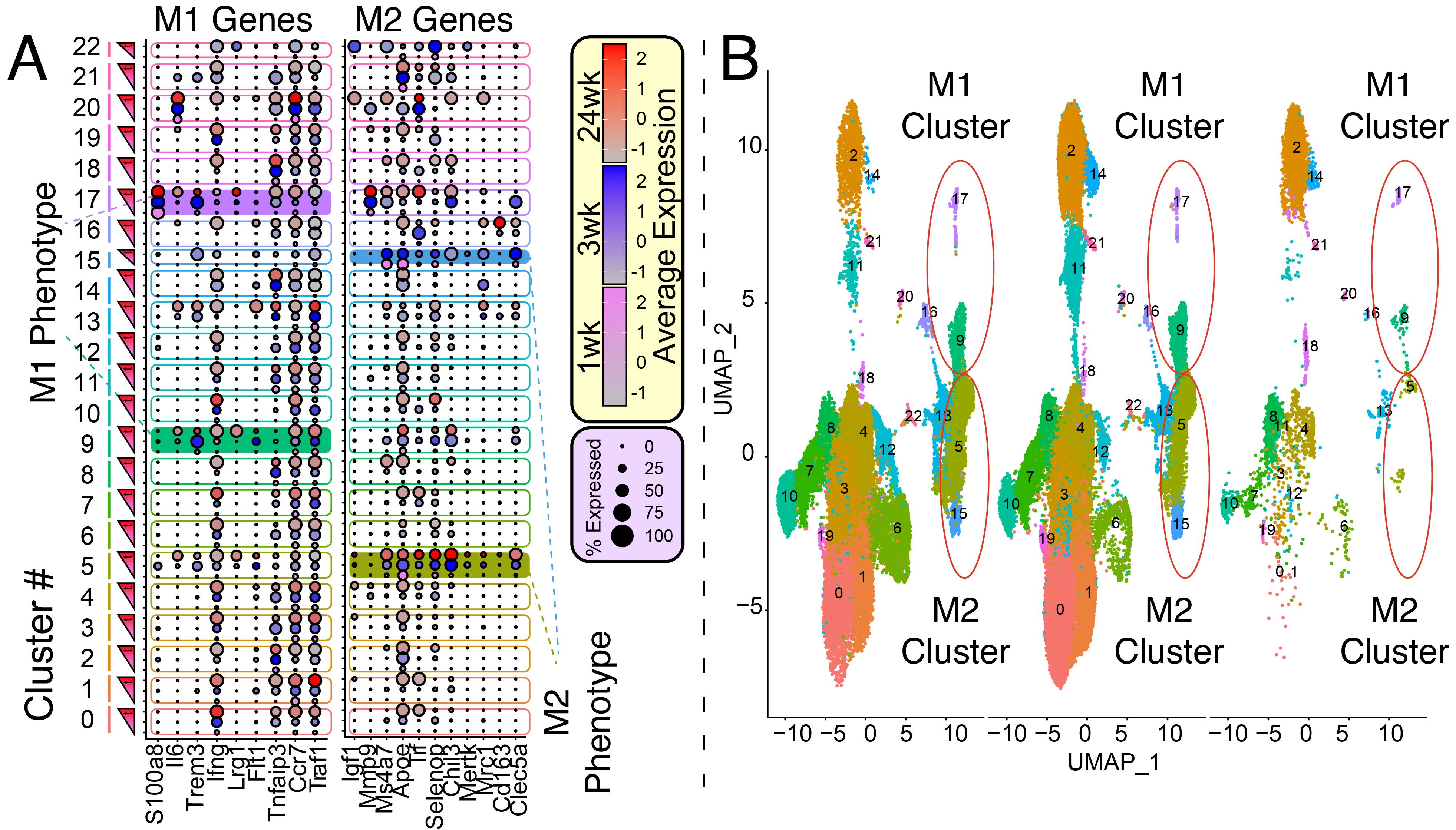
*Purpose: Kidney allografts transplanted across certain mouse strain combinations (e.g. DBA/2 to B6) are spontaneously accepted. We have recently shown that novel regulatory Tertiary Lymphoid Organs (rTLOs) form within accepted renal allografts and are made up of a diverse population of immune cells that aggregate within these structures. Here, we characterize the macrophage populations in rTLOs at the single-cell level.
*Methods: We performed life-sustaining DBA/2 to B6 kidney transplants. At 1 week (n=3), 3 weeks (n=5), and 24 weeks (n=3), transplanted kidneys were harvested, converted to single cells, sorted for CD45+, and analyzed by scRNAseq. The cDNA libraries underwent Next Generation Sequencing and the data was analyzed using the Seurat software package in RStudio. We present dot plots illustrating pro-inflammatory M1-like (S1008a, Il6, Trem3, Ifng, Lrg1, Flt1, Tnfaip3, Ccr7, Traf1) and anti-inflammatory/immunoregulatory M2-like (Igf1, Mmp9, Ms4a7, Apoe, Trf, Sepp1 [SELENOP], Chil3, Mertk, Mrc1, Cd163, Clec5a ) macrophage canonical genes across 22 UMAP clusters found in transplanted kidneys (Figure 1). Shaded-in clusters represent M1-like (9 and 17) and M2-like (5 and 15) populations in the data analysis. The size of the dot represents the frequency of gene expression in the cluster and color shade (e.g. red) represents the level of expression (e.g. darker = higher expression).
*Results: The macrophage signature appeared as early as 1-week post-transplantation. M1 gene expression levels plateaued and then decreased between 3 and 24 weeks post-transplantation, while M2 gene expression levels were amplified over time (clusters 5 and 15) and even began to be expressed within the M1 clusters (clusters 9 and 17) (Figure 1).
*Conclusions: Our gene expression data suggest that the macrophage signature in accepted kidneys transitioned from a pro-inflammatory, M1-like to an M2-like phenotype over time. The silencing of M1 macrophages and/or their conversion to M2 macrophages may be an important mechanism in the induction and maintenance of spontaneous allograft tolerance.
To cite this abstract in AMA style:
Guinn MT, Szuter ES, Yokose T, Ge J, Kwon B, Rosales I, Russell PS, Cuenca AG, Madsen JC, Colvin RB, Alessandrini A. Development of an M2 Macrophage Signature in Spontaneously Accepted Kidney Allografts [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/development-of-an-m2-macrophage-signature-in-spontaneously-accepted-kidney-allografts/. Accessed March 10, 2026.« Back to 2022 American Transplant Congress