Cellular Landscape of Steatotic Livers Revealed by Single-nuclei Rna Sequencing
1Surgery, James D Eason Transplant Institute, Memphis, TN, 2University of Maryland, Institute for Genome Sciences, Baltimore, MD, 3Program in Transplantation - University of Maryland, Baltimore, MD, 4Surgery, Division of Surgical Science, Baltimore, MD, 5Surgery, University of Maryland, Baltimore, MD, 6Surgery, Program in Transplantation - University of Maryland, Baltimore, MD
Meeting: 2021 American Transplant Congress
Abstract number: 356
Keywords: Genomic markers, Graft function, Liver, Obesity
Topic: Basic Science » Ischemia Reperfusion & Organ Rehabilitation
Session Information
Session Name: Ischemia Reperfusion & Organ Rehabilitation
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 8, 2021
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:30pm-6:35pm
Presentation Time: 6:30pm-6:35pm
Location: Virtual
*Purpose: Hepatic IRI represents a barrier to the use of available organs for transplant, particularly fatty livers. We aimed to discern the cellular subsets and individual transcriptomics that characterize human steatotic livers
*Methods: Single nuclei were isolated from normal (NL) and fatty (FL) (30% macrosteatosis) liver samples (n=4). Liver tissues were matched by age, sex, and gender. > 30,000 nuclei in Gel-Bead V3 were captured by using the droplet-based 10X Genomic Chromium Platform. Data was analyzed in CellRanger 3.1.0. Cell clustering using PCA followed by uniform manifold approximation and projection (UMAP) was done. Distinct clusters of cells based on GE of highly variable genes were identified. GE patterns between clusters were evaluated to find cluster-specific marker genes.
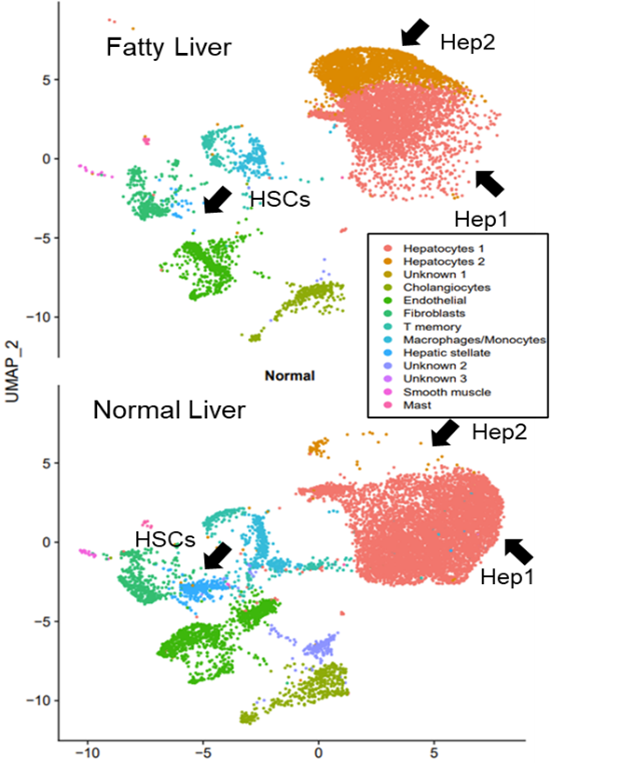
*Results: A total of 18 cell clusters were identified from the integration of total nuclei gene markers, which resulted in 13 cell clusters based on gene markers (representation of parenchymal and non-parenchymal liver cells) (Fig. 1). Normal to fatty liver comparisons were done and main observations included the identification of two hepatocyte cell subsets, Hep1 and Hep2; with Hep2 cells present predominantly in FL (Hep 2, NL= 87 vs. FL= 2390) and a significant decreased number of hepatic stellate cells (HSCs) in FL (NL= 345 vs. FL= 28) and endothelial cells (ECs) (N= 1122 vs. FL= 625) in FL. Analyses of DEGs between FL and NL Hep1 cells showed a lower expression of genes related to complement and coagulation components (CD46, CD55, C8, SERPINA1, fibrin, PLG, KNG1, KLKB1, KNG1) in FL Hep1. Hep2 cells were characterized by increased expression of genes related to acute phase response signaling, oxidation of lipids, FGF21 signaling, angiogenesis and development of vasculature. HSCs in FL presented an activated phenotype (while a quiescent HSC phenotype was observed in NL HSCs).
*Conclusions: These results support the use of single liver cell transcriptomics as a tool to identify specific cells/pathways that may represent targets for IRI therapeutic intervention for expanding the use of fatty liver organs
To cite this abstract in AMA style:
Kuscu C, Kuscu C, Shetty A, Bardhi E, Rousselle T, Eason J, Sraj M, Maluf D, Mas V. Cellular Landscape of Steatotic Livers Revealed by Single-nuclei Rna Sequencing [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/cellular-landscape-of-steatotic-livers-revealed-by-single-nuclei-rna-sequencing/. Accessed March 9, 2026.« Back to 2021 American Transplant Congress