A Comparison of Temporal Gene Expression Patterns in Kidney, Heart, Liver, and Lung Transplant Biopsies
1University of Alberta, Edmonton, AB, Canada, 2., ., AB, Canada
Meeting: 2020 American Transplant Congress
Abstract number: 312
Keywords: Gene expression, Heart, Kidney, Liver
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes III
Session Type: Oral Abstract Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:45pm
 Presentation Time: 4:27pm-4:39pm
Presentation Time: 4:27pm-4:39pm
Location: Virtual
*Purpose: Gene expression patterns change with time post-transplant. The MMDx system makes it possible to do cross-organ comparisons using the same microarray platform and gene sets. We compared these changes in kidney, heart, liver, and lung transplant biopsies, using gene sets representing rejection, injury, and atrophy-fibrosis.
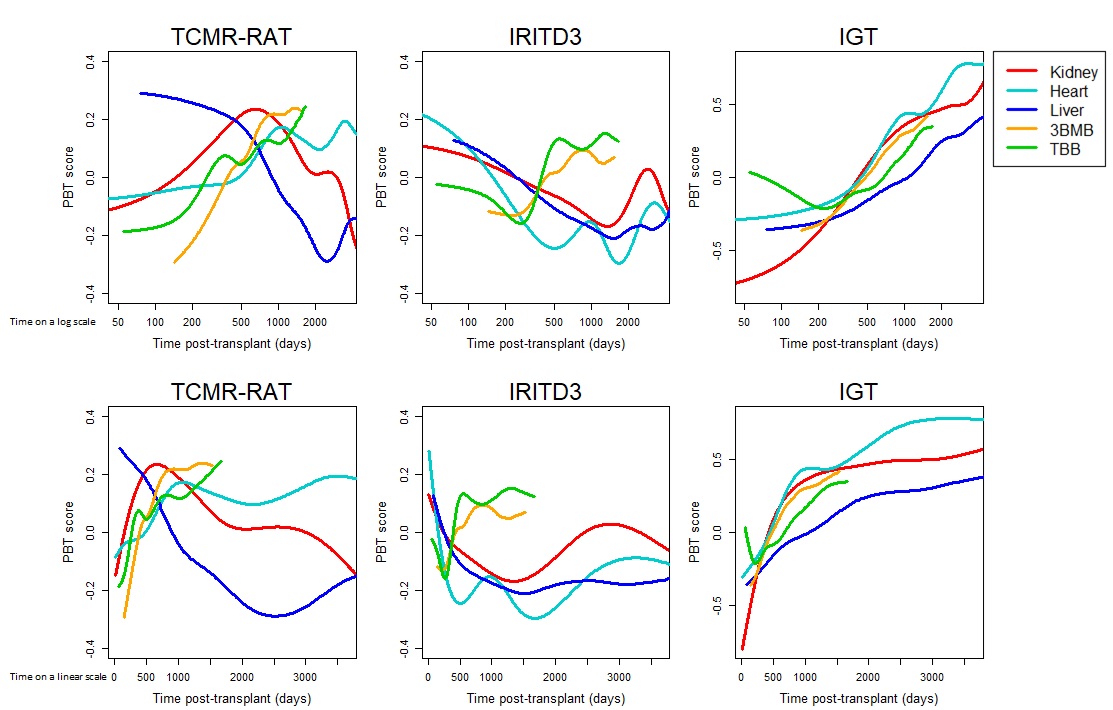
*Methods: Our populations consisted of 1526 kidney, 1320 heart, 337 liver, 457 transbronchial lung (TBB) and 314 mucosal lung (3BMB) biopsies. Various pathogenesis-based gene sets (PBTs) were examined – those shown in this abstract include one representing T cell-mediated rejection (TCMR-RAT), one for injury (IRITD3), and one for immunoglobulin genes (IGT). These PBTs were originally derived in human kidney transplants. Gene expression was assessed using Affymetrix PrimeView microarrays. PBT scores for each sample are calculated as the average fold change of all genes in the PBT vs the mean in a control population of relative normal transplant biopsies. Moving averages are based on consecutive sliding windows of 100 biopsies ordered by the time axis.
*Results: The results (Figure 1 – log time scale in the top row, linear time in the bottom row) indicate extensive inter-organ differences in rejection and injury, but consistency in the pattern of IGT expression. TCMR in livers (blue line) peaks early and becomes increasingly rare with time post-transplant, in keeping liver’s tolerogenic properties. In other organs, TCMR is delayed and peaks later, perhaps reflecting non-adherence. All organs experience early injury (IRITD3) that resolves, but lung TBB and 3BMB (green and orange) show a concerning late rise in IRITD3 after two years, accompanied by an increase in TCMR-RATs. However, all organs display a slow time-dependent increase in IGTs, as previously noted in kidneys, reflecting accumulation of plasma cells in areas of atrophy-scarring due to donation-implantation injury.
*Conclusions: Gene expression patterns exhibit characteristic changes that sometimes differ between the organs. The results show that cross-organ comparisons can provide useful insights into the biology of rejection, injury, and atrophy-scarring in organ transplant populations. The apparent instability of lung transplants compared to the stability of liver transplants represents an interesting correlation with clinical behaviors.
To cite this abstract in AMA style:
Halloran P, Reeve J. A Comparison of Temporal Gene Expression Patterns in Kidney, Heart, Liver, and Lung Transplant Biopsies [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/a-comparison-of-temporal-gene-expression-patterns-in-kidney-heart-liver-and-lung-transplant-biopsies/. Accessed February 21, 2026.« Back to 2020 American Transplant Congress