Common Molecular Features of Human Kidney Transplant Rejection Reflect Mechanistic Sharing Between T Cell-Mediated and Antibody-Mediated Rejection.
J. Venner,1,2 P. Halloran.2
1University of Queensland, Brisbane, Australia
2University of Australia, Edmonton, Canada.
Meeting: 2016 American Transplant Congress
Abstract number: 207
Keywords: Gene expression, Genomic markers, Graft-infiltrating lymphocytes, Inflammation
Session Information
Session Name: Concurrent Session: Kidney Immune Monitoring 2
Session Type: Concurrent Session
Date: Monday, June 13, 2016
Session Time: 2:30pm-4:00pm
 Presentation Time: 2:42pm-2:54pm
Presentation Time: 2:42pm-2:54pm
Location: Ballroom C
We previously reported the molecular features of human kidney transplant biopsies with T cell-mediated rejection (TCMR) (Am J Transplant 14(11):2565-76) and antibody-mediated rejection (ABMR) (Am J Transplant 15(5):1336-48), each compared to all other disease and injury states. The present study mapped the features shared by TCMR and ABMR, and sought to understand why this sharing was so extensive.
In 703 expression microarrays from prospectively collected kidney transplant indication biopsies (clinicaltrials.gov NCT01299168), we compared biopsies with rejection (TCMR, ABMR, mixed) to all others. Results in the Discovery Set (n=403) were highly conserved in the Validation Set (n=300) (Fig 1A-B). In the Combined Set (n=703), top rejection-associated transcripts were IFNG inducible molecules (e.g. CXCL9, 10, 11, PLA1A, GBP5), with P values as strong as 10-57 (Fig 1C). Molecules shared by NK and effector T cells were also strongly associated with rejection, including NK receptors NKG7, KLRK1 (NKG2D) and KLRD1 (CD94), CD8, and cytotoxic molecules PRF1, GNLY, and GZMA/B. Inflammasome transcripts were strongly associated with rejection (e.g. CASP1 and AIM2). However, many TCMR-selective molecules representing macrophages (e.g. ADAMDEC1) and costimulation (e.g. CTLA4, ICOS), as well as ABMR-selective molecules representing endothelium (e.g. DARC, VWF) and NK cells (e.g. SH2D1B) had reduced strengths when their association with rejection was considered (e.g. p values of 10-3).
The top rejection molecules reflected the similarity of the cognate events in T cell TCR signaling in TCMR and NK cell CD16a signaling in ABMR – e.g. IFNG release, reflected by IFNG inducible molecules – albeit localized in distinct compartments: interstitium in TCMR and microcirculation in ABMR. Combined with previous results in TCMR vs all other biopsies (T cell and DC/macrophage triggering) and ABMR vs all other biopsies (NK cells and endothelium), the rejection-associated transcripts provide a complete picture for both rejection phenotypes that could be useful for identifying new interventions against targets that could control both processes. 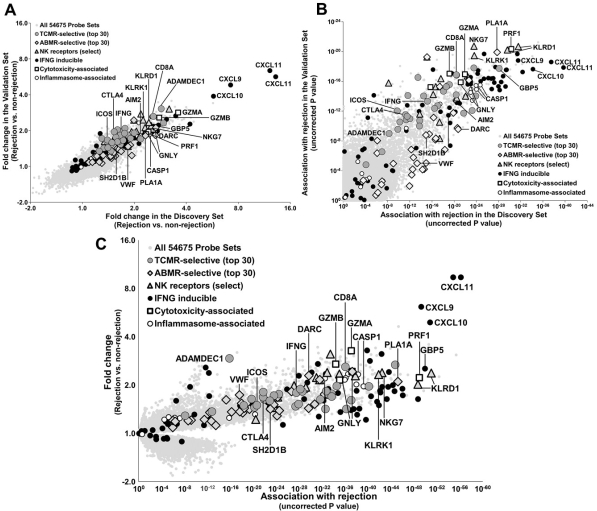
CITATION INFORMATION: Venner J, Halloran P. Common Molecular Features of Human Kidney Transplant Rejection Reflect Mechanistic Sharing Between T Cell-Mediated and Antibody-Mediated Rejection. Am J Transplant. 2016;16 (suppl 3).
To cite this abstract in AMA style:
Venner J, Halloran P. Common Molecular Features of Human Kidney Transplant Rejection Reflect Mechanistic Sharing Between T Cell-Mediated and Antibody-Mediated Rejection. [abstract]. Am J Transplant. 2016; 16 (suppl 3). https://atcmeetingabstracts.com/abstract/common-molecular-features-of-human-kidney-transplant-rejection-reflect-mechanistic-sharing-between-t-cell-mediated-and-antibody-mediated-rejection/. Accessed February 8, 2026.« Back to 2016 American Transplant Congress
