Donor-Derived Cell-Free DNA and Gene Expression Profiling for Surveillance of Renal Allograft Injury and Rejection
University of Maryland, Baltimore, MD
Meeting: 2022 American Transplant Congress
Abstract number: 1590
Keywords: Gene expression, Kidney, Rejection, Renal injury
Topic: Basic Science » Basic Clinical Science » 17 - Biomarkers: Clinical Outcomes
Session Information
Session Name: Biomarkers: Clinical Outcomes
Session Type: Poster Abstract
Date: Tuesday, June 7, 2022
Session Time: 7:00pm-8:00pm
 Presentation Time: 7:00pm-8:00pm
Presentation Time: 7:00pm-8:00pm
Location: Hynes Halls C & D
*Purpose: Donor derived cell free DNA (dd-cfDNA) and gene expression profiling offer non-invasive methods of early detection of graft injury and rejection in solid organ transplant recipients. These biomarkers are orthogonal since they evaluate very different aspects of kidney and immune responses to injury events. In addition to detection of rejection, dd-cfDNA and gene expression profiles may be reliable markers of other clinical events leading to graft injury. We hypothesized that use of these tests together would successfully identify patients who have a clinically significant event inform clinicians of the need for additional investigations, such as imaging, blood/urine tests, or kidney biopsy.
*Methods: A sample of 41 patients from our institution who were enrolled in the Outcomes of Kidneycare on Renal Allografts (OKRA) Study were identified to evaluate the efficacy of dd-cfDNA (using AlloSure Kidney) and gene expression profiles (using AlloMap Kidney), together with serum creatinine and urine microalbumin, to identify rejection or other allograft injury without invasive testing. Descriptive statistics were used in addition to data obtained in for-cause and surveillance biopsies.
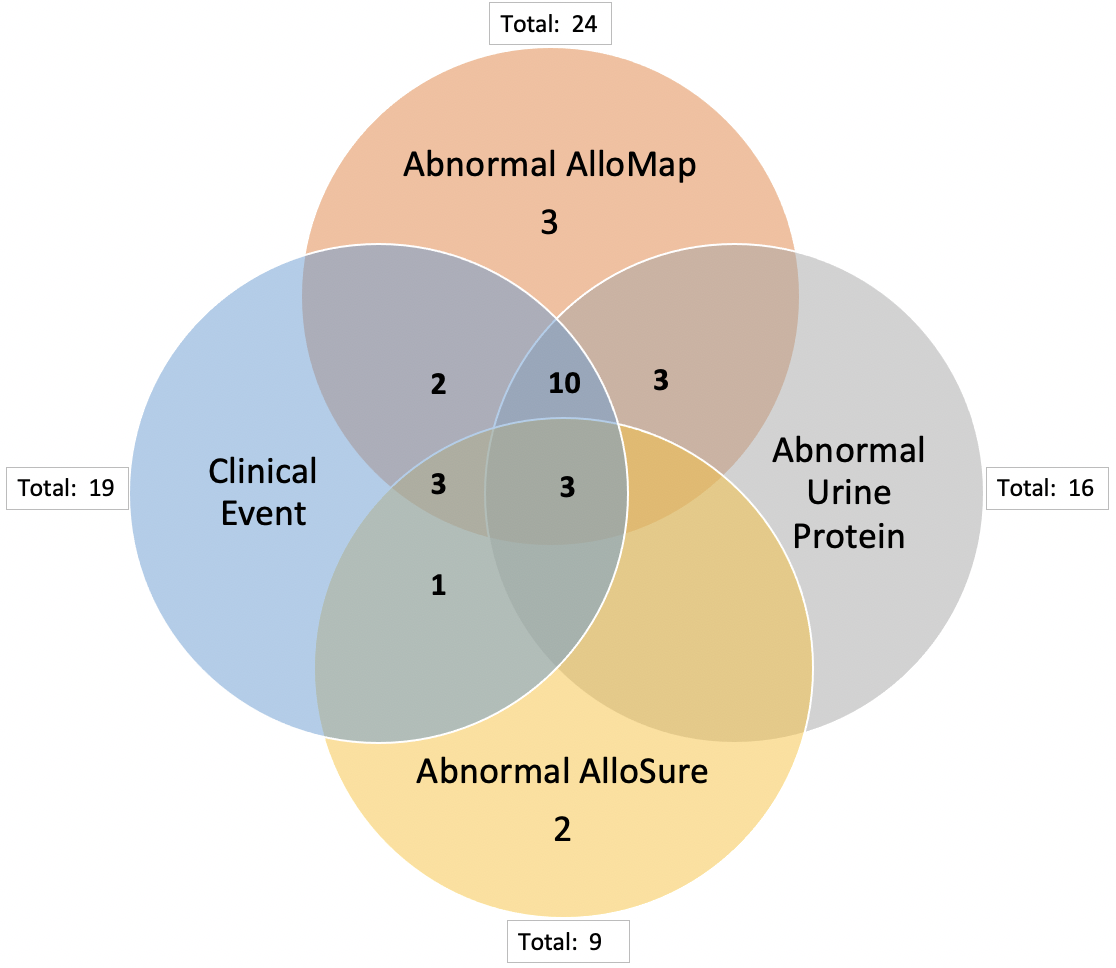
*Results: From this sample, 26 patients had at least one abnormal dd-cfDNA test (defined as an AlloSure Kidney > 1.00%, nine patients total) or gene expression profile (defined as AlloMap Kidney > 10, 24 patients total). Serum creatinine was elevated in 25 patients with an abnormal AlloSure Kidney or AlloMap Kidney test. An abnormal dd-cfDNA or gene expression profile test successfully identified 19 patients who were confirmed to have a combination of allograft rejection (8 patients), BK viremia (4 patients), tacrolimus toxicity (6 patients), and/or recurrent FSGS (2 patients) on for-cause and surveillance biopsies. Two additional patients were noted to have an infection. Only thirteen patients with a clinical event also had abnormal urine microalbumin.
*Conclusions: Dd-cfDNA and gene expression profiling can be used as orthogonal tests to analyze different aspects of graft injury and rejection. The novel use of both dd-cfDNA and gene-expression profiling in conjunction with serum creatinine and urinalysis can improve early detection of significant graft injury, including rejection, BK viremia, tacrolimus toxicity, and recurrent FSGS, through non-invasive means. Larger populations will be needed to determine the specificity and sensitivity of these tests in combination for each clinical scenario.
To cite this abstract in AMA style:
Carrier AN, Crane A, Pinedo M, Bartosic A, Haririan A, Scalea JR, Weir MR, Bromberg JS. Donor-Derived Cell-Free DNA and Gene Expression Profiling for Surveillance of Renal Allograft Injury and Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/donor-derived-cell-free-dna-and-gene-expression-profiling-for-surveillance-of-renal-allograft-injury-and-rejection/. Accessed February 2, 2026.« Back to 2022 American Transplant Congress