Leveraging Single-Cell Signatures in Kidney Transplant Biopsies Uncovers Immunological Heterogeneity Within Rejection Phenotypes
1Microbiology, Immunology and Transplantation, KU Leuven, Leuven, Belgium, 2Department of Morphology and Molecular Pathology, University Hospitals Leuven, Leuven, Belgium, 3Faculty of Medicine, KU Leuven, Leuven, Belgium, 4Laboratory for Translational Genetics, Department of Human Genetics, KU Leuven, Leuven, Belgium, 5Necker-Enfants Malades Institute, Institut national de la santé et de la recherche médicale (Inserm), Université de Paris, Paris, France, 6ESAT Stadius Center for Dynamical Systems, Signal Processing and Data Analytics, KU Leuven, Leuven, Belgium
Meeting: 2022 American Transplant Congress
Abstract number: 626
Keywords: Biopsy, Gene expression, Kidney transplantation, Rejection
Topic: Basic Science » Basic Science » 02 - Acute Rejection
Session Information
Session Time: 5:30pm-7:00pm
 Presentation Time: 5:30pm-7:00pm
Presentation Time: 5:30pm-7:00pm
Location: Hynes Halls C & D
*Purpose: Rejection of the kidney allograft is hallmarked by the accumulation of inflammatory cells within the renal architecture, leading to microvascular or tubulointerstitial inflammation. However, our knowledge of the immune cell composition and its spatial organization in rejection remains limited.
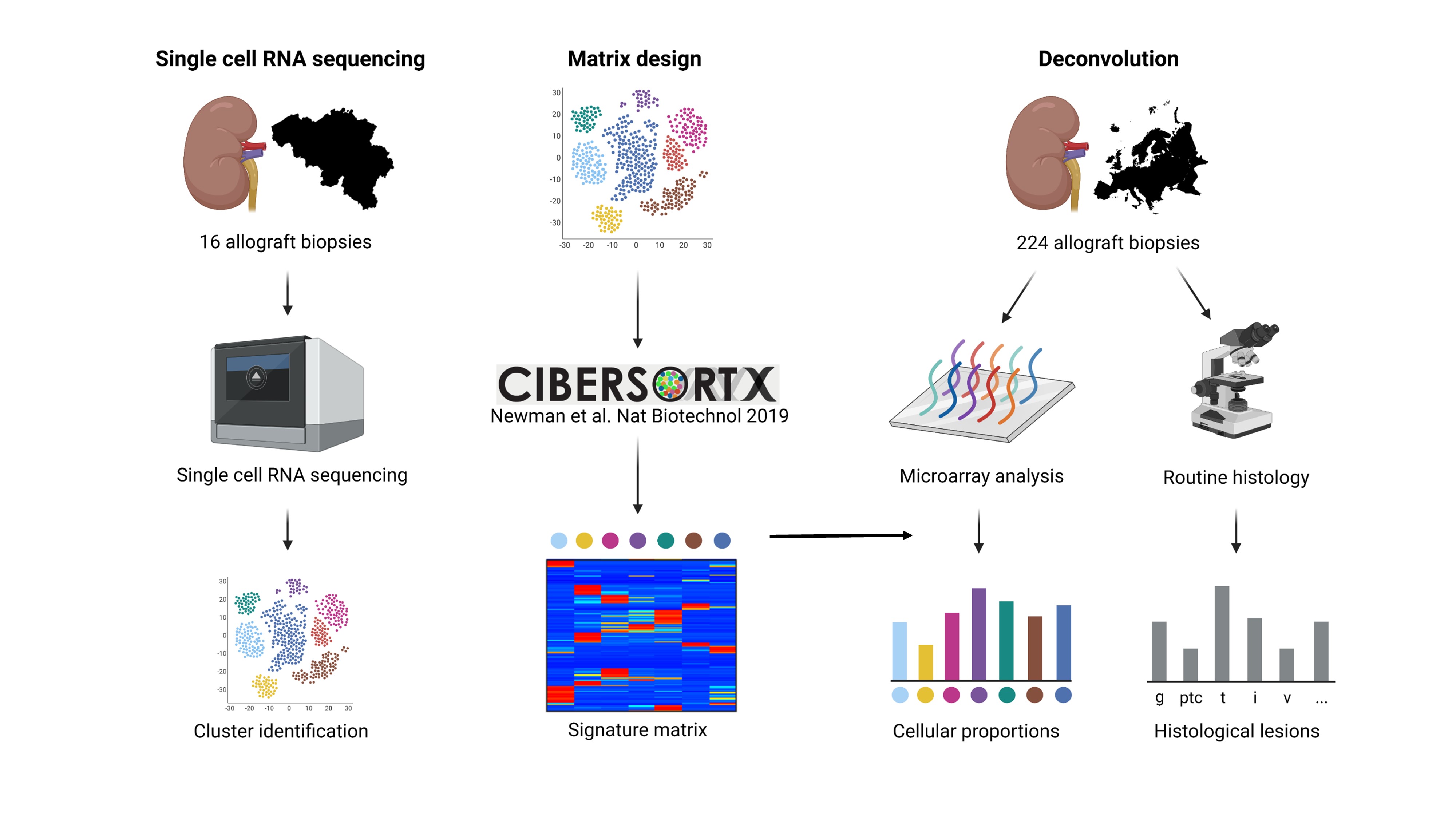
*Methods: Droplet-based single-cell RNA sequencing of 16 kidney transplant biopsies was performed, yielding 35,152 single cell transcriptomes. Cell-type specific gene expression signatures were constructed using CIBERSORTx, and applied for deconvolution of the cellular fractions in an independent microarray cohort of 224 kidney transplant biopsies (Figure). Multiplex immunofluorescence analysis of 86 biopsy slides was applied to unravel the spatial distribution of macrophages, T cells and NK cells in the allograft.
*Results: Single-cell RNA sequencing identified 18 distinct clusters, corresponding to epithelial, endothelial, stromal and immune cell subsets on the basis of canonical expression markers. Cellular deconvolution of biopsies in the microarray cohort and multiplex imaging revealed a specific association between intravascular NK cell accumulation and vascular lesions, whereas CD8+ T cells, monocytes and macrophages were involved in both vascular and tubulointerstitial lesions. However, across histological patterns of inflammation, important variation was found in the cellular composition of the immune infiltrate.
*Conclusions: Single-cell derived signatures of the human kidney transplant can be applied in bulk transcriptomics to analyze the immune cell repertoire in rejection. Although specific leukocyte types are related to microvascular and tubulointerstitial inflammation, important cellular heterogeneity was found in biopsies with similar light microscopic appearances, highlighting the need to better understand the cellular ecosystems of rejection.
To cite this abstract in AMA style:
Callemeyn J, Lamarthée B, Koshy P, Debyser T, Loon EVan, Franken A, Brussel TVan, Arijs I, Lambrechts D, Rabant M, Senev A, Vaulet T, Tinel C, Naesens M. Leveraging Single-Cell Signatures in Kidney Transplant Biopsies Uncovers Immunological Heterogeneity Within Rejection Phenotypes [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/leveraging-single-cell-signatures-in-kidney-transplant-biopsies-uncovers-immunological-heterogeneity-within-rejection-phenotypes/. Accessed March 3, 2026.« Back to 2022 American Transplant Congress