Molecular Classifiers for Chronic Lung Allograft Dysfunction Transbronchial and Mucosal Biopsies Predict Clinical CLAD and Graft Loss
M. D. Parkes1, P. F. Halloran2, K. M. Halloran3, &. the INTERLUNG Study Group4
1Alberta Transplant Applied Genomics Centre, Edmonton AB, AB, Canada, 2Alberta Transplant Applied Genomics Centre, Edmonton, AB, Canada, 3University of Alberta, Edmonton AB, AB, Canada, 4., ., AB, Canada
Meeting: 2021 American Transplant Congress
Abstract number: 92
Keywords: Gene expression, Graft survival, Lung transplantation, Risk factors
Topic: Clinical Science » Lung » Lung: All Topics
Session Information
Session Name: How to Expect the Unexpected- Incorporating Predictors into Lung Transplant Decision Making
Session Type: Rapid Fire Oral Abstract
Date: Saturday, June 5, 2021
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:05pm-6:10pm
Presentation Time: 6:05pm-6:10pm
Location: Virtual
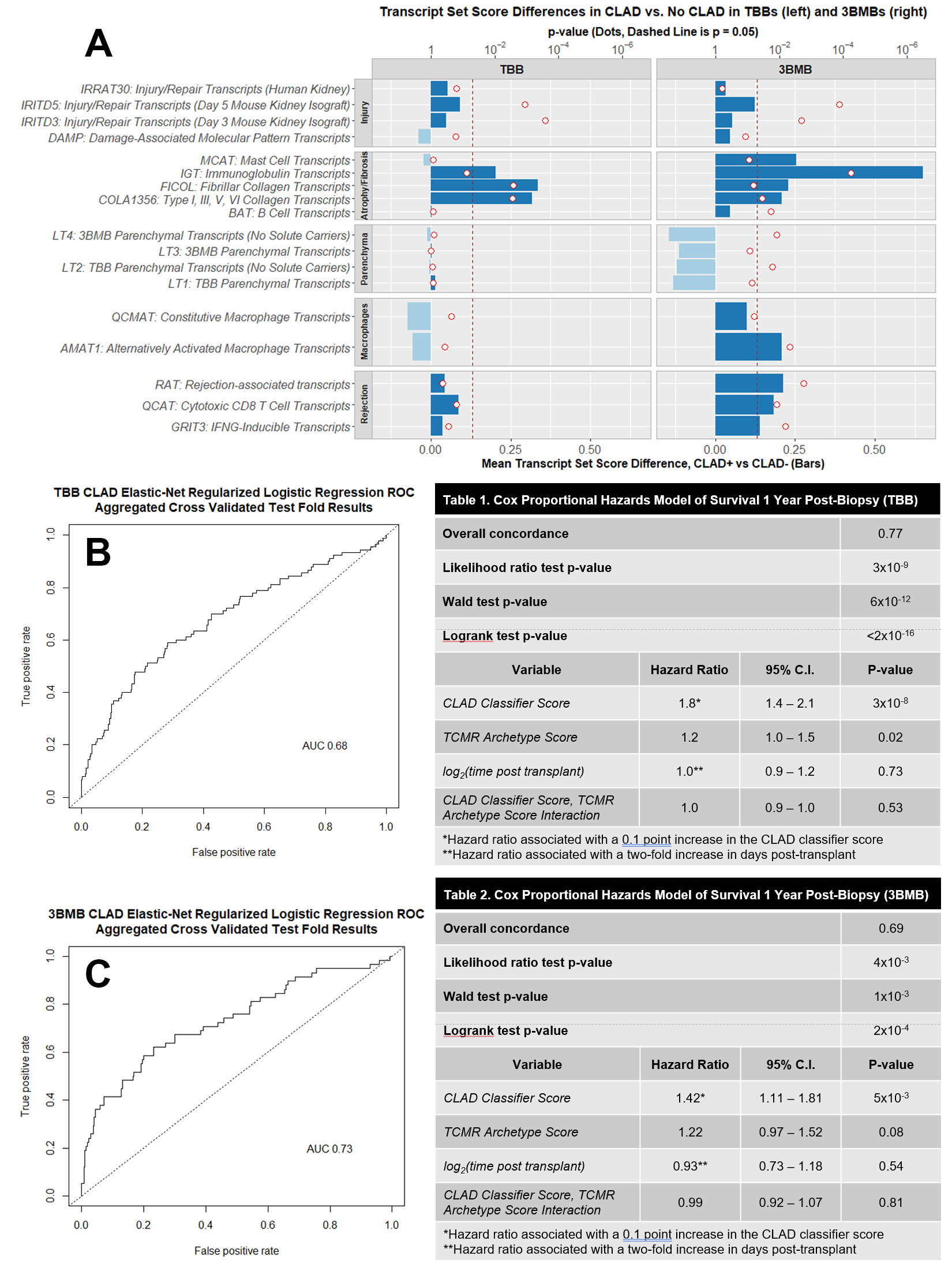
*Purpose: Chronic lung allograft dysfunction (CLAD) portends poor outcomes yet is poorly understood. In the multi-center INTERLUNG study we measured transcripts in transbronchial biopsies (TBBs) and mucosal biopsies from the third bronchial bifurcation (3BMBs) using microarrays and documented CLAD-associated gene expression. We studied whether probabilistic gene-based estimates of CLAD are associated with risk of graft loss by 1 year post-biopsy.
*Methods: We collected 498 TBBs from 423 transplants (3BMB: 324, 285) with known CLAD status. We identified CLAD-associated transcripts and gene set scores. Elastic net -regularized logistic regressions predicting CLAD were fit on 20 genes associated with CLAD after correcting for clinical confounders, time post-transplant, and MMDx T cell mediated rejection (TCMR). We fit multivariate Cox proportional hazards models on predicted CLAD probability, MMDx TCMR probability, time post-transplant, and interaction between CLAD and MMDx TCMR probabilities. TBBs represented 35 failures, 3BMBs 27.
*Results: CLAD-associated gene expression changes reflected parenchymal dedifferentiation and response-to-wounding, especially atrophy/fibrosis, but some were also associated with clinical confounders and, in 3BMBs, time post-transplant. Some IFNG-inducible genes (e.g. HLA-DOA) were associated with CLAD in 3BMBs but not TBBs. CLAD and DSA were uncorrelated.
The regressions predicted CLAD with AUC 0.68 (TBB) and 0.73 (3BMB). In TBBs each 0.1 increase in CLAD probability was associated with 1.8x hazard of graft loss by 1 year post-biopsy (p=3×10-8), compared to 1.4x in 3BMBs (p=0.005). The CLAD probability outperformed the clinical diagnosis in TBBs although not in 3BMBs. Each 0.1 increase in MMDx TCMR probability multiplied hazard by 1.2 in TBBs (p=0.02) and 3BMBs (p=0.08), but it did not compound hazards with the CLAD score.
*Conclusions: CLAD resembles a parenchymal response to wounding that overlaps other pathologies and is better captured in 3BMBs. The CLAD classifiers agreed more closely with clinical diagnoses in 3BMBs than TBBs, but in both cases the scores reflected a CLAD-like phenotype and were associated with increased risk of graft loss by 1 year post-biopsy. Molecular detection of CLAD-like changes may aid patient management when other conditions exclude CLAD or when the diagnosis is uncertain.
To cite this abstract in AMA style:
Parkes MD, Halloran PF, Halloran KM. Molecular Classifiers for Chronic Lung Allograft Dysfunction Transbronchial and Mucosal Biopsies Predict Clinical CLAD and Graft Loss [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/molecular-classifiers-for-chronic-lung-allograft-dysfunction-transbronchial-and-mucosal-biopsies-predict-clinical-clad-and-graft-loss/. Accessed February 21, 2026.« Back to 2021 American Transplant Congress