Transcriptomic Profiling of Paired Serum and Biopsy Samples from FSGS Patients
1Transplant Research Institute, James D. Eason Transplant Institute, Department of Surgery, University of Tennessee Health Science Center, Memphis, TN, 2Department of Systems and Computational Biology, University of Hyderabad, Hyderabad, India, 3Department of Medicine and Surgery, Albert Einstein College of Medicine, New York, NY
Meeting: 2020 American Transplant Congress
Abstract number: D-329
Keywords: Gene expression, Genomics, Kidney, Renal dysfunction
Session Information
Session Name: Poster Session D: B-cell / Antibody /Autoimmunity
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Pathogenesis and molecular footprints of focal segmental glomerulosclerosis (FSGS) after kidney transplantation whether it is primary or secondary is unknown. Herein, we studied a small cohort of patients from whom we collected kidney biopsies and serum at the time of diagnosis.
*Methods: To investigate the molecular characteristics of FSGS, we performed gene expression analysis using Affymetrix microarray from kidney biopsies and cell free small noncoding RNA profiling using Next Generation sequencing from serum in patients with primary or secondary FSGS.
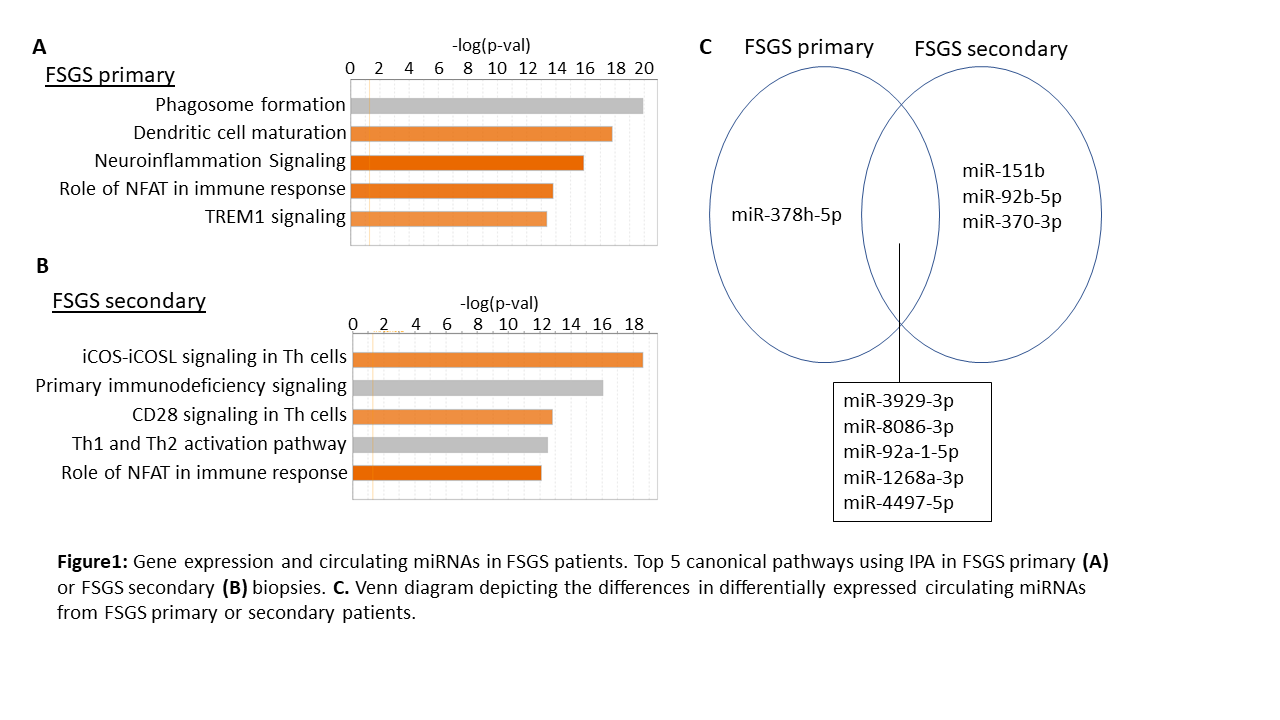
*Results: Analysis of altered canonical pathways in FSGS primary or secondary using Ingenuity Pathway Analysis (IPA) showed a lot of similarities between FSGS primary and secondary. On the other hand, there are couple of pathways that are specific to FSGS types: TREM1 signaling in FSGS primary and primary immunodeficiency signaling in FSGS secondary (Fig 1A). Even though FSGS primary and FSGS secondary share differentially expressed circulating miRNAs, there are specific ones for FSGS primary (e.g. miR-378h) and secondary (e.g. miR-151b, miR-92b, miR-370). Integration of miRNA and gene expression profiles using Ingenuity Pathway Analysis (IPA) suggested that decrease in miR-92a levels may result in upregulation of genes that are important for B cell development and activation.
*Conclusions: Integration of circulating miRNA and gene expression profiles from FSGS patients will shed more light in the disease pathogenesis and lead to new therapeutics. In the past, higher number of B cells have been detected in pediatric FSGS patients who were later treated with Rituximab. Taken together, our results suggest that role of B cells in FSGS needs to be explored carefully.
To cite this abstract in AMA style:
Kuscu C, Kiran M, Wolen A, Kuscu C, Bajwa A, Eason J, Maluf D, Akalin E, Mas V. Transcriptomic Profiling of Paired Serum and Biopsy Samples from FSGS Patients [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/transcriptomic-profiling-of-paired-serum-and-biopsy-samples-from-fsgs-patients/. Accessed February 19, 2026.« Back to 2020 American Transplant Congress