miRNA Signatures as Prediction Tool for DSA Development and Rejection
1Terasaki Research Institute, Los Angeles, CA, 2East Carolina University, Greenville, NC
Meeting: 2019 American Transplant Congress
Abstract number: B36
Keywords: Genomic markers, Graft survival, Kidney transplantation, Non-invasive diagnosis
Session Information
Session Name: Poster Session B: Biomarkers, Immune Monitoring and Outcomes
Session Type: Poster Session
Date: Sunday, June 2, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
*Purpose: Early and convenient diagnosis is urgently needed for acute and chronic transplant rejection due to its high risk of the loss of the transplant. An ongoing chronic or acute inflammation is associated in allograft rejection and is going along with the development of donor-specific antibodies (DSA). The discovery of novel classes of non-coding RNAs (ncRNAs) in many biofluids like serum, plasma and urine has revolutionized medicine. Circulating microRNAs (miRNAs) as a part of ncRNAs are promising biomarkers of inflammation and the deeper understanding of the mechanisms underlying the development of DSA, however, there is still little known about circulating miRNAs involved in controlling inflammation and DSA. In this study we analyzed DSA+ and DSA- serum samples to identify a specific signature of miRNAs that will help to predict DSA development and rejection.
*Methods: We performed an enriched small RNA preparation from kidney transplant patients (15 DSA+, 5 DSA- and 5 healthy controls). The enriched RNA was reverse transcribed and a screening for miRNA was conducted using a human miRNA RT-qPCR screen platform (Exiqon human panel 1 + 2). The expression of the miRNAs was normalized to the expression profile of miR-16-5p. For the analysis of the data we used the software platform GenEx and the pathway analysis software IPA.
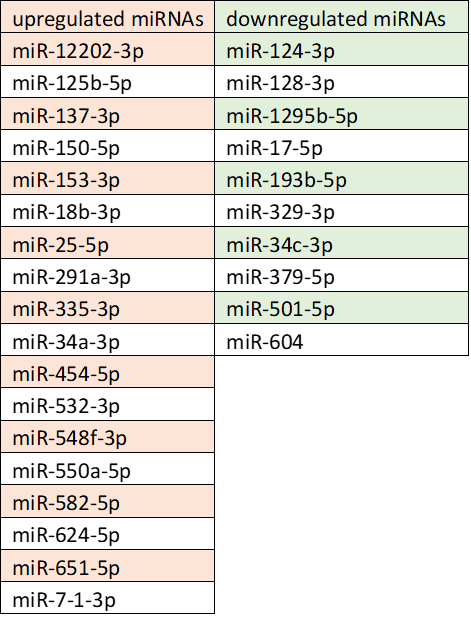
*Results: When we compare DSA- vs DSA+ samples we were able to identify 10 down and 18 upregulated miRNAs with a p value of <0.05. The pathway analysis of this molecules shows that they are related to organismal injury and two of the miRNAs especially described in the literature as renal inflammation and nephritis related.
*Conclusions: In this study we were able to identify a panel of miRNAs that plays a role in the regulation of cellular stress and renal inflammation/nephritis in DSA+ patients. We are currently getting a deeper look into this panel to define a diagnostic panel and to validate is relevance. To identify missing or unknown miRNAs and to validate our first results we analyze RNA isolated from exosomes at the timepoint of DSA onset with microRNA NGS as a new and a powerful tool to identify and quantitatively decode the entire population of microRNAs in a second set of patient samples.
To cite this abstract in AMA style:
Hamdorf M, Rebellato LM, Briley KP, Everly MJ. miRNA Signatures as Prediction Tool for DSA Development and Rejection [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/mirna-signatures-as-prediction-tool-for-dsa-development-and-rejection/. Accessed February 18, 2026.« Back to 2019 American Transplant Congress