Validation of CYP4F11 as a Marker of Accommodation in Biopsies from Recipients of an ABO-Incompatible Renal Transplant.
1Centre for Complement and Inflammation Research, Imperial College, London, United Kingdom
2King Saud University, Riyadh, Saudi Arabia
3Imperial College Healthcare NHS Trust, London, United Kingdom
Meeting: 2017 American Transplant Congress
Abstract number: A123
Keywords: Gene expression, Genomic markers, Kidney transplantation, Tolerance
Session Information
Session Name: Poster Session A: Diagnostics/Biomarkers Session I
Session Type: Poster Session
Date: Saturday, April 29, 2017
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall D1
Recipients of an ABO incompatible transplant have circulating anti-AB antibodies, A and/or B antigens expressed on endothelium and usually C4d deposition on the endothelium, yet with adequate immunosuppressive treatment, this seldom leads to antibody-mediated rejection. Previous work using RNA sequencing compared ABO-incompatible (ABOi) and ABO-compatible (ABOc) surveillance biopsies with normal histology and identified a number of differentially expressed genes. Here we validate CYP4F11 as a marker of accommodation in an independent set of biopsies.
RNA was obtained from formalin fixed paraffin embedded tissue from 13 ABOc and 7 ABOi renal transplant patients. Samples were a mix of indication and surveillance biopsies but none had histological evidence of rejection. Eighteen genes considered the top hits in sequencing analysis were assayed using a Nanostring nCounter custom panel.
Comparison between two groups demonstrated a reduced expression of CYP4F11 in ABOi patients compared to ABOc (p=0.0063, Mann-Whitney test), confirming association with accommodation of the renal allograft. 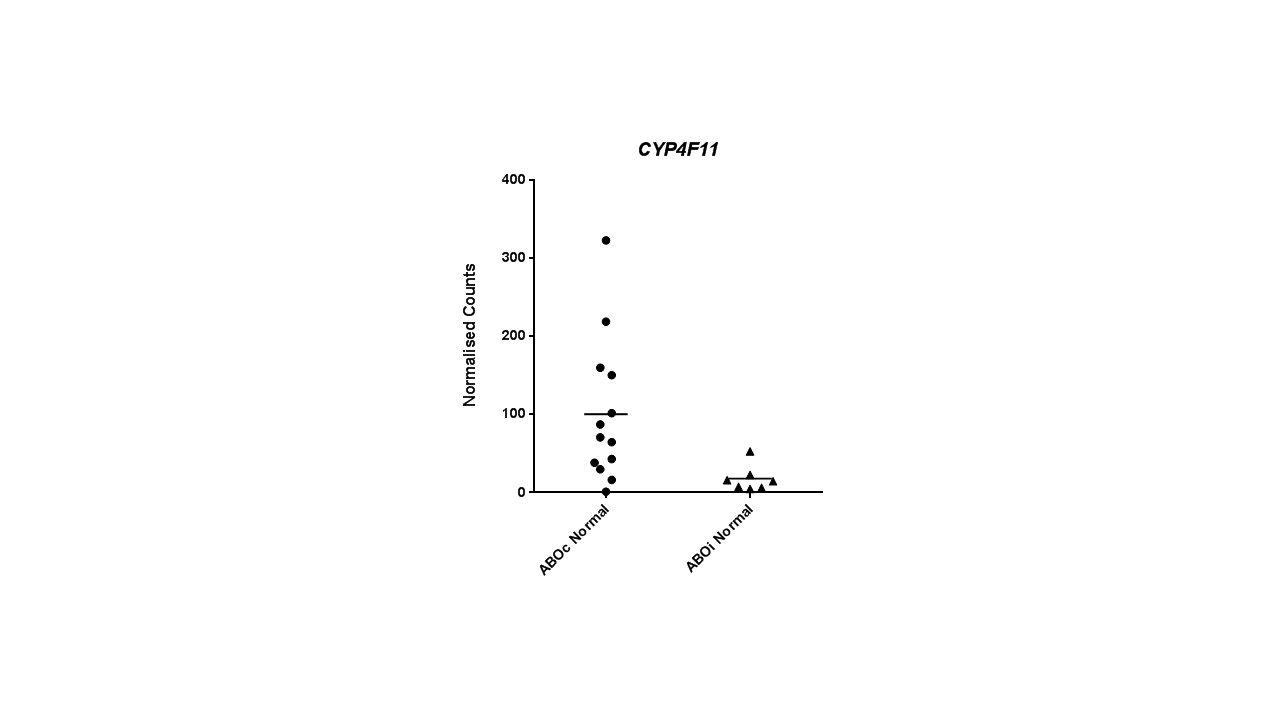 None of the other genes demonstrated a significant differential expression between the two groups. The study may be under-powered to validate any of the other genes with individual effects of genes potentially very small, rendering detection difficult. Additional sample numbers may assist in validating or disproving associations.
None of the other genes demonstrated a significant differential expression between the two groups. The study may be under-powered to validate any of the other genes with individual effects of genes potentially very small, rendering detection difficult. Additional sample numbers may assist in validating or disproving associations.
Most of the substrates of cytochrome p450 4F isoforms are eicosanoids, which play important roles in the inflammatory response. Cytochrome 4F enzymes metabolize arachidonic acid to 20-hydroxyeicosatetraenoic acid (20-HETE). CYP4F11 is known to have high expression in the kidney and 20-HETE inhibitors have demonstrated improved renal microvascular function in hypertension. The down-regulation of CYP4F11 in ABOi transplants may be likewise protecting the microvasculature.
CITATION INFORMATION: Dominy K, Willicombe M, Al Johani T, Galliford J, McLean A, Cook T, Roufosse C. Validation of CYP4F11 as a Marker of Accommodation in Biopsies from Recipients of an ABO-Incompatible Renal Transplant. Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Dominy K, Willicombe M, Johani TAl, Galliford J, McLean A, Cook T, Roufosse C. Validation of CYP4F11 as a Marker of Accommodation in Biopsies from Recipients of an ABO-Incompatible Renal Transplant. [abstract]. Am J Transplant. 2017; 17 (suppl 3). https://atcmeetingabstracts.com/abstract/validation-of-cyp4f11-as-a-marker-of-accommodation-in-biopsies-from-recipients-of-an-abo-incompatible-renal-transplant/. Accessed February 18, 2026.« Back to 2017 American Transplant Congress
