Validation Of The Optimized Prediction Of Mortality (Opom) Machine Learning Model For Non-hepatocellular Carcinoma Candidates Awaiting Liver Transplant
1Multi-Organ Transplant, Department of Surgery, University of Toronto, Toronto, ON, Canada, 2Mechanical and Industrial Engineering, University of Toronto, Toronto, ON, Canada
Meeting: 2019 American Transplant Congress
Abstract number: C288
Keywords: Allocation, Liver transplantation, Methodology, Resource utilization
Session Information
Session Name: Poster Session C: Liver: MELD, Allocation and Donor Issues (DCD/ECD)
Session Type: Poster Session
Date: Monday, June 3, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
Session Information
Session Name: Young Investigator Oral Communication Session
Session Type: Concurrent Session
Date: Monday, June 3, 2019
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:30pm-4:40pm
Presentation Time: 4:30pm-4:40pm
Location: Room 208
*Purpose: Currently the Model for End-Stage Liver Disease (MELD) is used to rank candidates for liver transplant in many jurisdictions. However, MELD does not provide equitable organ allocation to all waitlisted candidates. Recently, the Optimized Prediction of Mortality (OPOM) model was developed to predict 3-month waitlist mortality or drop-out, calibrated on the SRTR dataset. OPOM is an Optimal Classification Tree machine learning model that considers numerous dynamic variables and was previously shown to be superior to MELD-based allocation using simulation models. It is uncertain whether this model is valid in other patient populations.
*Methods: From 2003-2018, 3142 non-HCC patients were waitlisted and 2056 patients were transplanted at the University of Toronto. This time-stamped dataset prospectively captured all candidate details used in the OPOM. Each valid candidate timepoint was evaluated using the previously calibrated classification tree to categorize 3-month drop-out status. Data was coded into the 5th layer of OPOM with 32 leaf nodes. Using a rounding threshold of 0.5, the classification of Toronto data was compared to the risk prediction of OPOM. The AUC of the OPOM was also calculated and compared to MELD and MELD-Na.
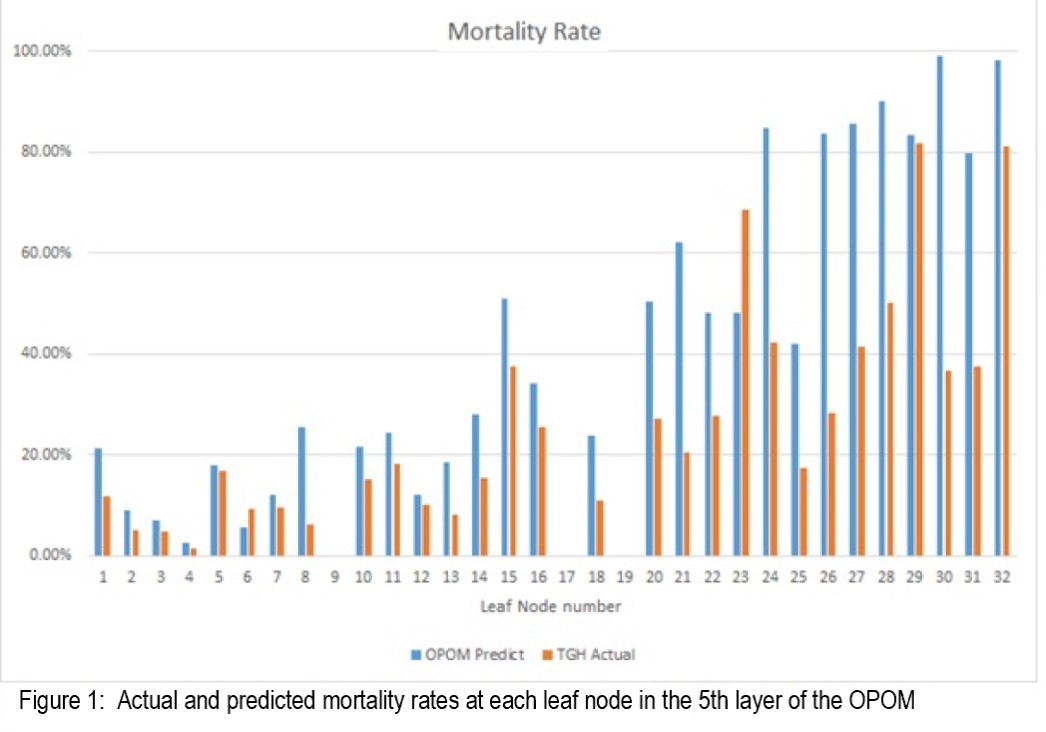
*Results: A total of 22,111 discrete candidate timepoints in 2376 evaluable patients were assessed. The differences between the actual and predicted outcomes at the OPOM leaf nodes were assessed (Figure 1).
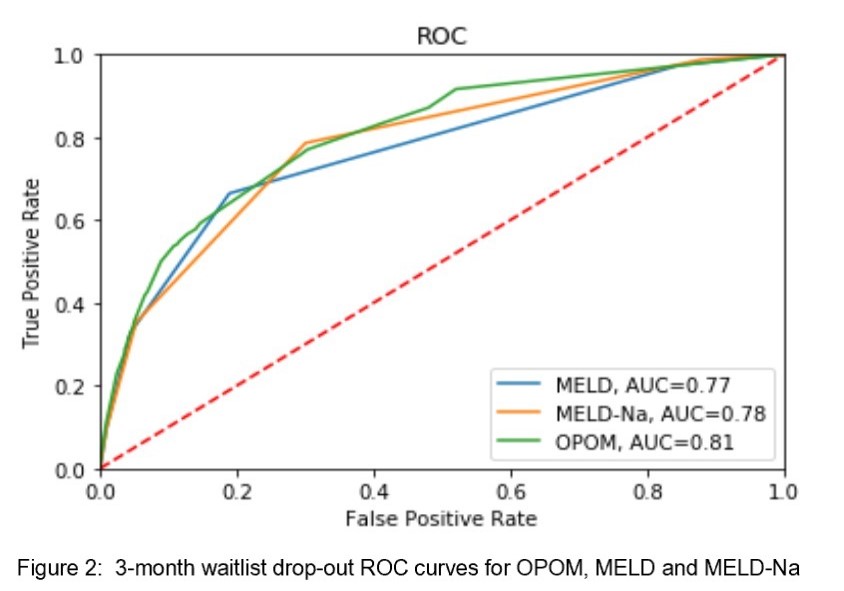
The predictive accuracy is 90.0% with a positive predictive value of 39.6% and a sensitivity of 34.6%. The OPOM had an AUC of 0.81 for 3-month drop-out, which was superior to MELD (0.77) and MELD-Na (0.78)(Figure 2).
*Conclusions: The OPOM model was validated on a large cohort of Canadian liver transplant candidates. These results suggest that a machine learning approach may be valuable in stratifying the sickest patients for future transplant waitlist prioritization.
To cite this abstract in AMA style:
Lau L, Xie X, Jia F, Greig PD, Chan TC. Validation Of The Optimized Prediction Of Mortality (Opom) Machine Learning Model For Non-hepatocellular Carcinoma Candidates Awaiting Liver Transplant [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/validation-of-the-optimized-prediction-of-mortality-opom-machine-learning-model-for-non-hepatocellular-carcinoma-candidates-awaiting-liver-transplant/. Accessed March 9, 2026.« Back to 2019 American Transplant Congress