Validation of a Novel Gene Expression Biomarker of Rejection Following Liver Transplantation
1Gastroenterology & Hepatology
Comprehensive Transplant Center, Northwestern University, Chicago, IL, 2University of California San Diego, San Diego, CA, 3Scripps Clinic, La Jolla, CA, 4University of Arizona College of Medicine Tucson, Tucson, AZ
Meeting: 2021 American Transplant Congress
Abstract number: 118
Keywords: Genomic markers, Immunosuppression, Liver transplantation, Rejection
Topic: Clinical Science » Biomarkers, Immune Assessment and Clinical Outcomes
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes - II
Session Type: Rapid Fire Oral Abstract
Date: Sunday, June 6, 2021
Session Time: 4:30pm-5:30pm
 Presentation Time: 5:05pm-5:10pm
Presentation Time: 5:05pm-5:10pm
Location: Virtual
*Purpose: Non-invasive biomarkers distinguishing early signs of immune activation vs. quiescence could more objectively guide immunosuppression management after liver transplantation (LT). Our aim was to validate blood-based gene expression profiles that can serially detect immune activation prior to acute rejection (AR) distinct from immune quiescence (non-AR).
*Methods: Gene expression results in LT recipients with AR vs. non-AR (combination of other causes of graft dysfunction and normal graft function) were analyzed from two published cohorts (Northwestern University (NU); multicenter NIAID CTOT-14). A 70:30 approach (61 AR; 162 non-AR) was used for discovery and validation. Serial sample results before AR and non-AR were available in the CTOT-14 study (all <1 year post-LT). Microarray data was normalized utilizing a liver-specific fRMA vector. Variable selection was done using a specific variance (0.5), expression (8.0) and bootstrap (0.3) threshold.
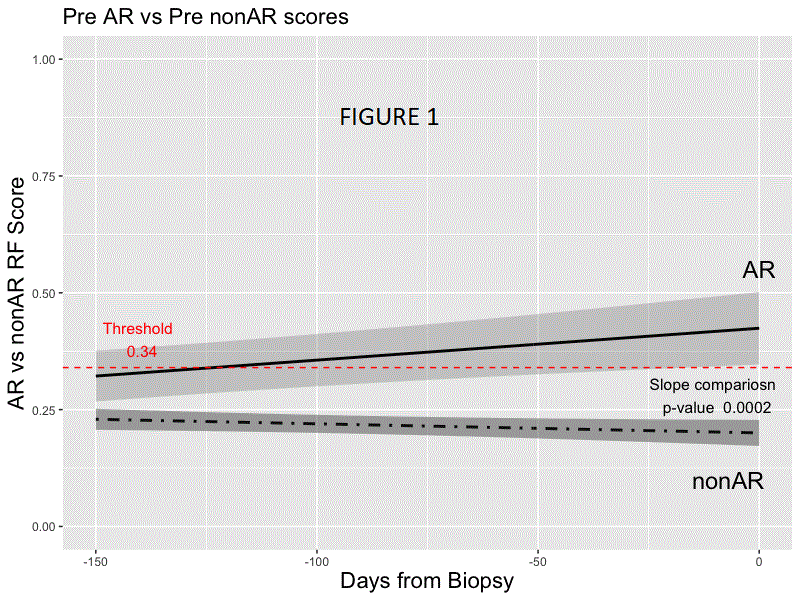
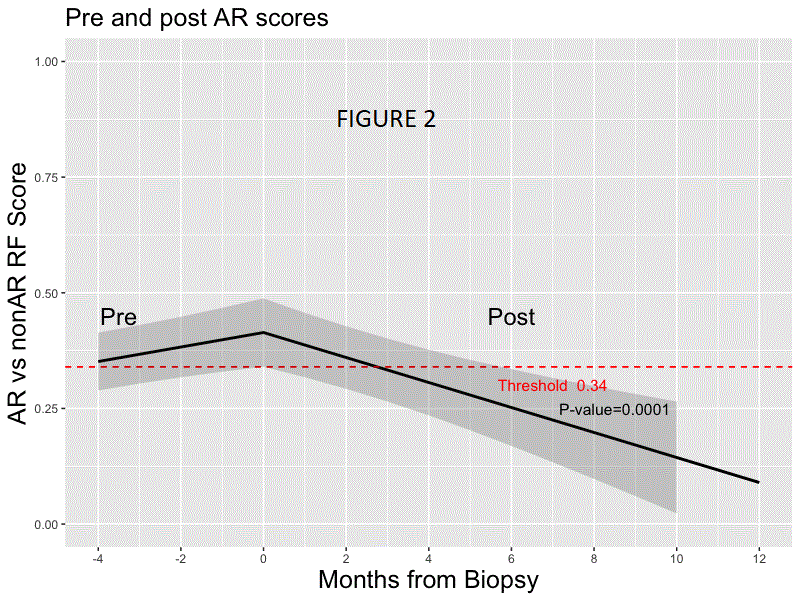
*Results: Random forest modeling was used to generate a classifier on the discovery set distinguishing AR vs. non-AR (AUC 0.83; accuracy of 0.78, sensitivity 0.70, specificity 0.81, PPV 0.54, NPV 0.89; F-score .61). The final 59 probe model and locked probability threshold performed well on the validation set (accuracy of 0.72, sensitivity 0.67, specificity 0.73, PPV 0.48, NPV 0.86; F-score 0.56). The probability score line slopes were positive preceding AR and negative preceding non-AR and following AR treatment (all p<0.001; Figs. 1 & 2) in CTOT-14; the threshold was crossed at ~120 days prior to AR. Among the top 36 pathways that were significant using IPA (p <0.01) for the 59 probes, >40% were within immune response pathways, including allograft rejection.
*Conclusions: We have developed a blood-based biologically relevant genomic biomarker diagnostic for AR that can be detected prior to AR-associated graft injury and distinct from non-AR. Prospective interventional trials are needed to evaluate its utility in precision-guided immunosuppression following LT.
To cite this abstract in AMA style:
Levitsky J, Kandpal M, Whisenant T, Guo K, Kurian S, Abecassis M. Validation of a Novel Gene Expression Biomarker of Rejection Following Liver Transplantation [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/validation-of-a-novel-gene-expression-biomarker-of-rejection-following-liver-transplantation/. Accessed February 25, 2026.« Back to 2021 American Transplant Congress