Using a Unique Repository of Pancreas Transplant Biopsies to Validate the Tissue Common Response Module (tCRM) Score as a Tool to Assess the Severity of Pancreas Transplant Rejection
1University of California, San Francisco, San Francisco, CA, 2University of Maryland Medical Center, Baltimore, MD
Meeting: 2022 American Transplant Congress
Abstract number: 430
Keywords: Biopsy, Genomic markers, Pancreas transplantation, Rejection
Topic: Clinical Science » Pancreas » 65 - Pancreas and Islet: All Topics
Session Information
Session Name: Pancreas and Islet: All Topics
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 7, 2022
Session Time: 3:30pm-5:00pm
 Presentation Time: 4:00pm-4:10pm
Presentation Time: 4:00pm-4:10pm
Location: Hynes Room 210
*Purpose: Analysis of transcriptional data from kidney, heart, liver, and lung transplant rejection led to development of the tissue Common Response Module (tCRM) score based on 11 genes (BASP1, ISG20, PSMB9, RUNX3, TAP1, NKG7, LCK, INPP5D, CXCL9, CD6, CXCL10) which can diagnose the severity of rejection, but have not been validated in pancreas transplant (PT). We aimed to use a unique repository of PT biopsies to evaluate the locked tCRM score and identify additional markers of PT acute rejection (AR).
*Methods: We performed a retrospective study applying gene expression analysis to RNA isolated from formalin-fixed paraffin-embedded tissue from 51 pancreas biopsies, grouped by grade of acute cellular rejection (Grade 1, 2, and 3) versus normal. Gene expression analysis was performed using Nanostring Cancer Immune v1.1 oligonucleotide set of 804 unique genes. Differential gene expression analysis was performed as well as calculation of the tCRM score and pathway analysis to evaluate biological significance.
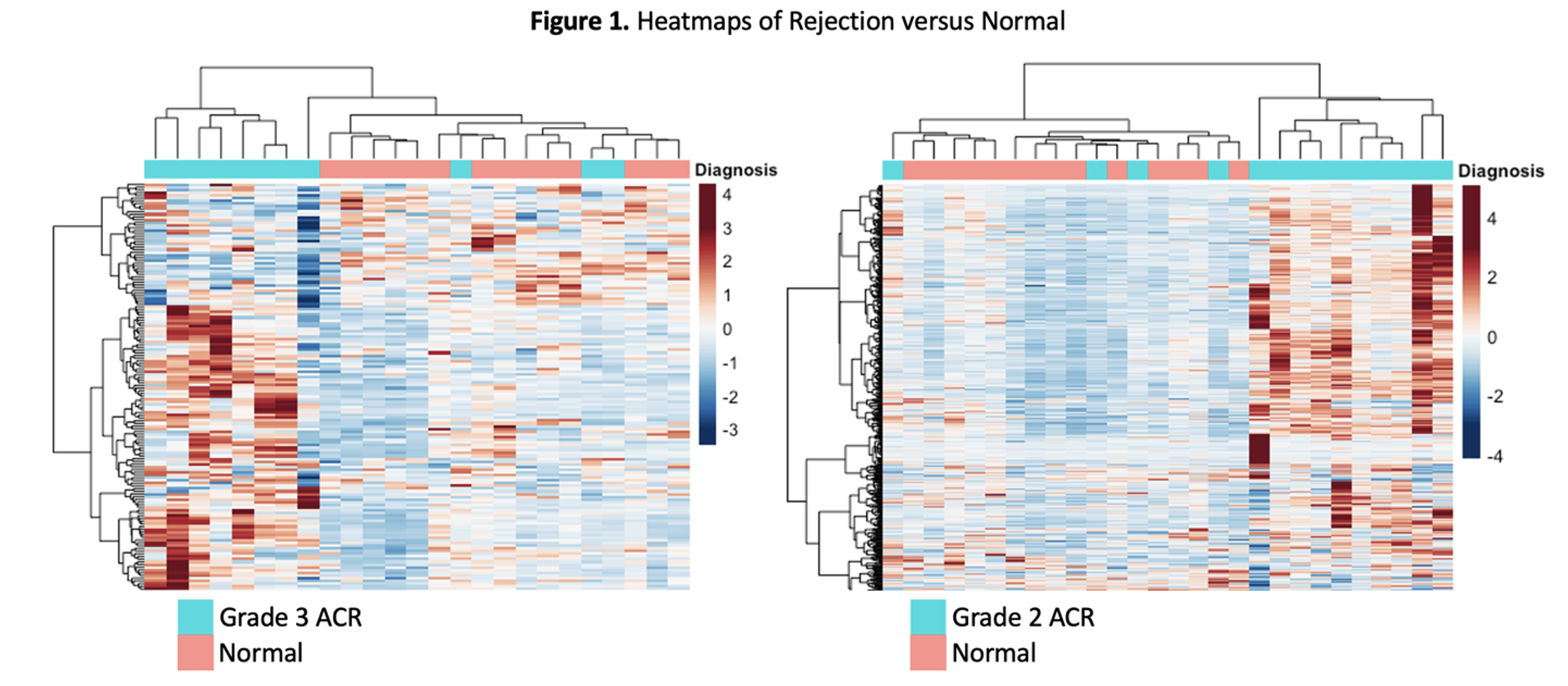
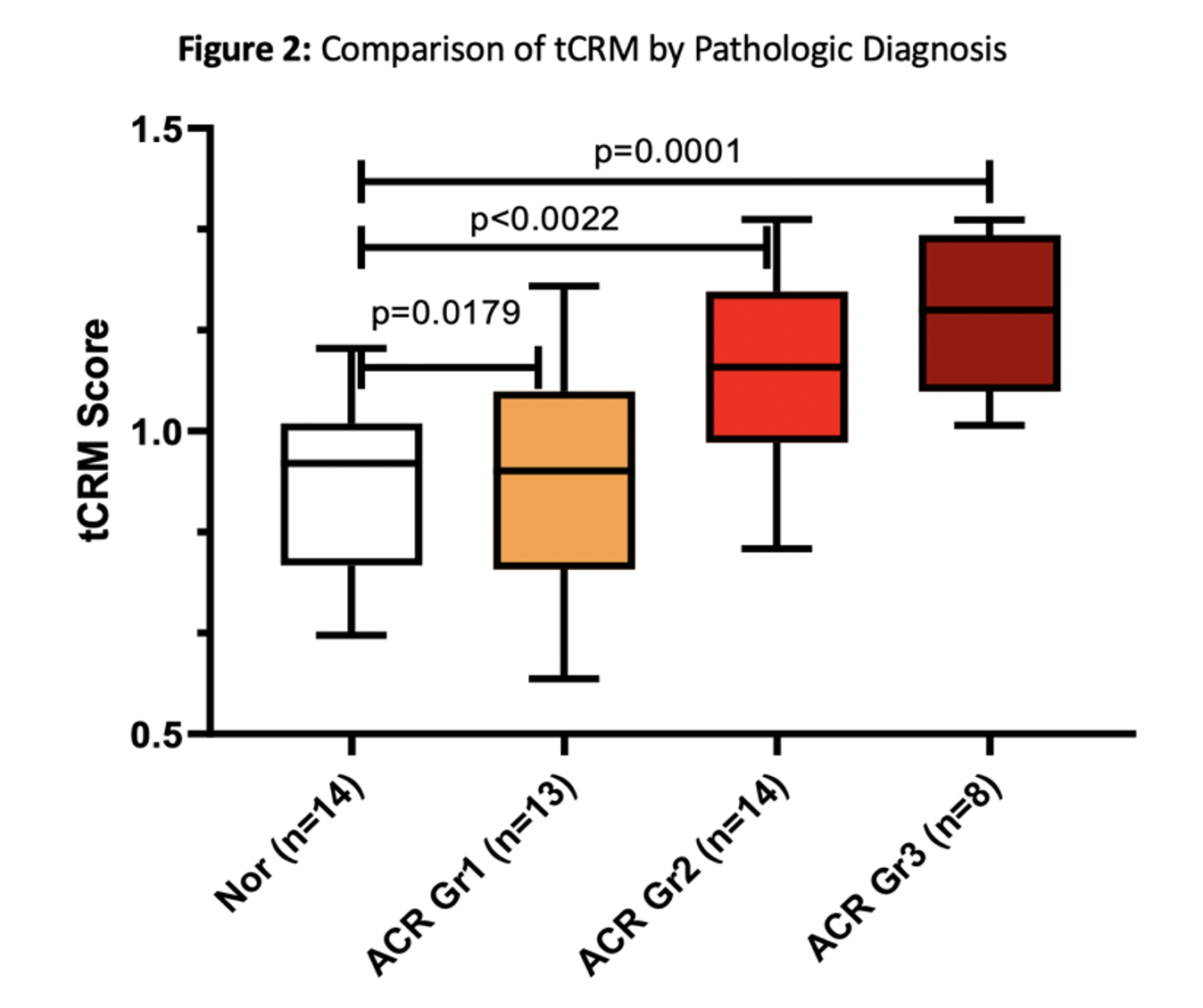
*Results: Significant differences were seen with higher grades of rejection among several transcripts (Fig.1). Of the 22 genes differentially expressed in Grade 3 ACR, 18 were also differentially expressed in Grade 2 ACR. The tCRM score captures the same message with a scaled score that increases across rejection grades (Fig.2). Within the 11 tCRM genes diagnostic of rejection in other solid organ transplants, 9 were significant for PT rejection, with the last two genes (CXCL9 and CXCL10) approaching significance (p=0.055 and 0.08 respectively).
*Conclusions: The bulk of AR signal in PT, similar to other solid organs, is attributable to infiltration by activated leukocytes, with background tissue injury specific transcriptional changes. A common hub of immune mediated genes highly significant in rejection (tCRM) and not confounded by tissue source (kidney, heart, liver and lung), were also highly informative for diagnosis and quantification of AR injury in PT. This presents the potential to monitor for rejection using this panel of genes in patients.
To cite this abstract in AMA style:
Kelly Y, Zarinsefat A, Meier R, Sarwal M, Stock P, Laszik Z, Sigdel T. Using a Unique Repository of Pancreas Transplant Biopsies to Validate the Tissue Common Response Module (tCRM) Score as a Tool to Assess the Severity of Pancreas Transplant Rejection [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/using-a-unique-repository-of-pancreas-transplant-biopsies-to-validate-the-tissue-common-response-module-tcrm-score-as-a-tool-to-assess-the-severity-of-pancreas-transplant-rejection/. Accessed February 21, 2026.« Back to 2022 American Transplant Congress