Unbiased Sequencing of Urine Cell-Free DNA (cfDNA) to Evaluation Urine Virome and Associated Renal Allograft Damage
1Transplantation Medicine-Nephrology, Weill Cornell Medicine, NY, NY
2Biomedical Engineering, Cornell University, Ithaca, NY.
Meeting: 2018 American Transplant Congress
Abstract number: 498
Keywords: Graft function, Kidney transplantation, Monitoring, Polyma virus
Session Information
Session Name: Concurrent Session: Biomarkers, Immune Monitoring and Outcomes: Clinical
Session Type: Concurrent Session
Date: Tuesday, June 5, 2018
Session Time: 4:30pm-6:00pm
 Presentation Time: 5:18pm-5:30pm
Presentation Time: 5:18pm-5:30pm
Location: Room 602/603/604
Intro: Virus reactivation leading to graft damage is a frequent post-kidney transplant (KTx) complication. Immunosuppressed individuals are at risk for multiple virus infections simultaneously. Current clinical practice is to use separate urine/blood samples for detection of each virus individually. A single assay that detects the entire urine virome as well as allograft damage would be of value.
Methods: cfDND was extracted from 1ml urine supernatants collected at time of BKVN biopsy (n=24) and in a control cohort, 10 stable KTx with normal histology + 27 with UTI (no biopsy), and sequenced on Illumina NextSeq platform using single-stranded library preparation. Sequences reads were aligned with the human reference genome UCSC hg19 and the unaligned reads were extracted and mapped to viral reference genomes. cfDNA donor fraction (DF) was used in gender-mismatched recipients to assess graft damage.
Results: Urine cfDNA detected viral copies in 100% of BKVN patients, in 50% with stable function and 22% with UTIs (Fig1).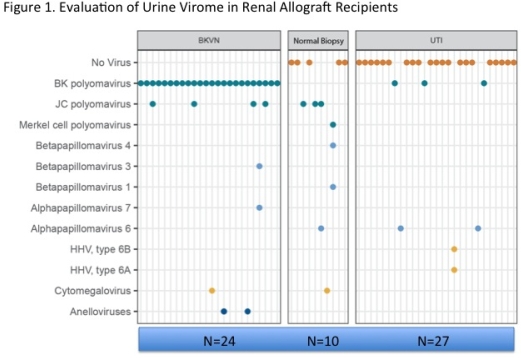 Multiple viral genomes were more frequent in BKVN (33%) vs. non-BKVN (7.4%) cohort. Presence of BKV sequences correlated with BKVN (P<0.001) & relative genomic abundance of BKV correlated with absolute copies of BKV VP1 mRNA levels (rs=0.72;P<0.001). DF was higher in BKVN compared to control cohort (Fig2). DF positively correlated with same day creatinine levels (rs=0.37;P=0.04).
Multiple viral genomes were more frequent in BKVN (33%) vs. non-BKVN (7.4%) cohort. Presence of BKV sequences correlated with BKVN (P<0.001) & relative genomic abundance of BKV correlated with absolute copies of BKV VP1 mRNA levels (rs=0.72;P<0.001). DF was higher in BKVN compared to control cohort (Fig2). DF positively correlated with same day creatinine levels (rs=0.37;P=0.04). 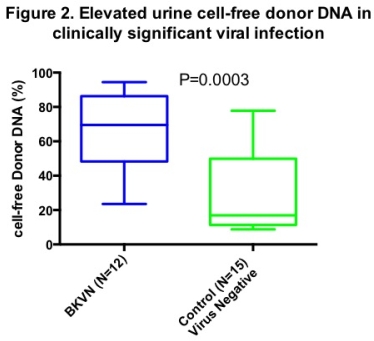
Conclusion: Our data demonstrate the feasibility & utility of cfDNA to measure urine virome comprehensively. Our novel assay offers a noninvasive measure of viral burden and allograft damage. Future studies will apply this novel technology to monitor KTx recipients & personalize management.
CITATION INFORMATION: Dadhania D., Burnham P., Lee J., Snopkowski C., Li C., Albakry S., Yang H., Muthukumar T., De Vlaminck I., Suthanthiran M. Unbiased Sequencing of Urine Cell-Free DNA (cfDNA) to Evaluation Urine Virome and Associated Renal Allograft Damage Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Dadhania D, Burnham P, Lee J, Snopkowski C, Li C, Albakry S, Yang H, Muthukumar T, Vlaminck IDe, Suthanthiran M. Unbiased Sequencing of Urine Cell-Free DNA (cfDNA) to Evaluation Urine Virome and Associated Renal Allograft Damage [abstract]. https://atcmeetingabstracts.com/abstract/unbiased-sequencing-of-urine-cell-free-dna-cfdna-to-evaluation-urine-virome-and-associated-renal-allograft-damage/. Accessed March 2, 2026.« Back to 2018 American Transplant Congress
