The Study on the DNA Methylation Dynamic Changes after Renal Transplantation
Nephrology, The First Affiliated Hospital of Zhejiang University, Hangzhou, China
Meeting: 2019 American Transplant Congress
Abstract number: C48
Keywords: Genomics, Graft failure
Session Information
Session Name: Poster Session C: Kidney Complications: Late Graft Failure
Session Type: Poster Session
Date: Monday, June 3, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
*Purpose: Kidney transplantation is the most effective therapeutic regimen for most ESRD patients. However, graft dysfunction remains to be a major challenge. DNA methylation is critical for regulating the transcriptional expression and determines the fate of many cells, including immune cells and fibroblasts. Given that epigenetics is an emerging area in organ transplantation, this study aims to describe the pattern of DNA methylation between allograft function and dysfunction.
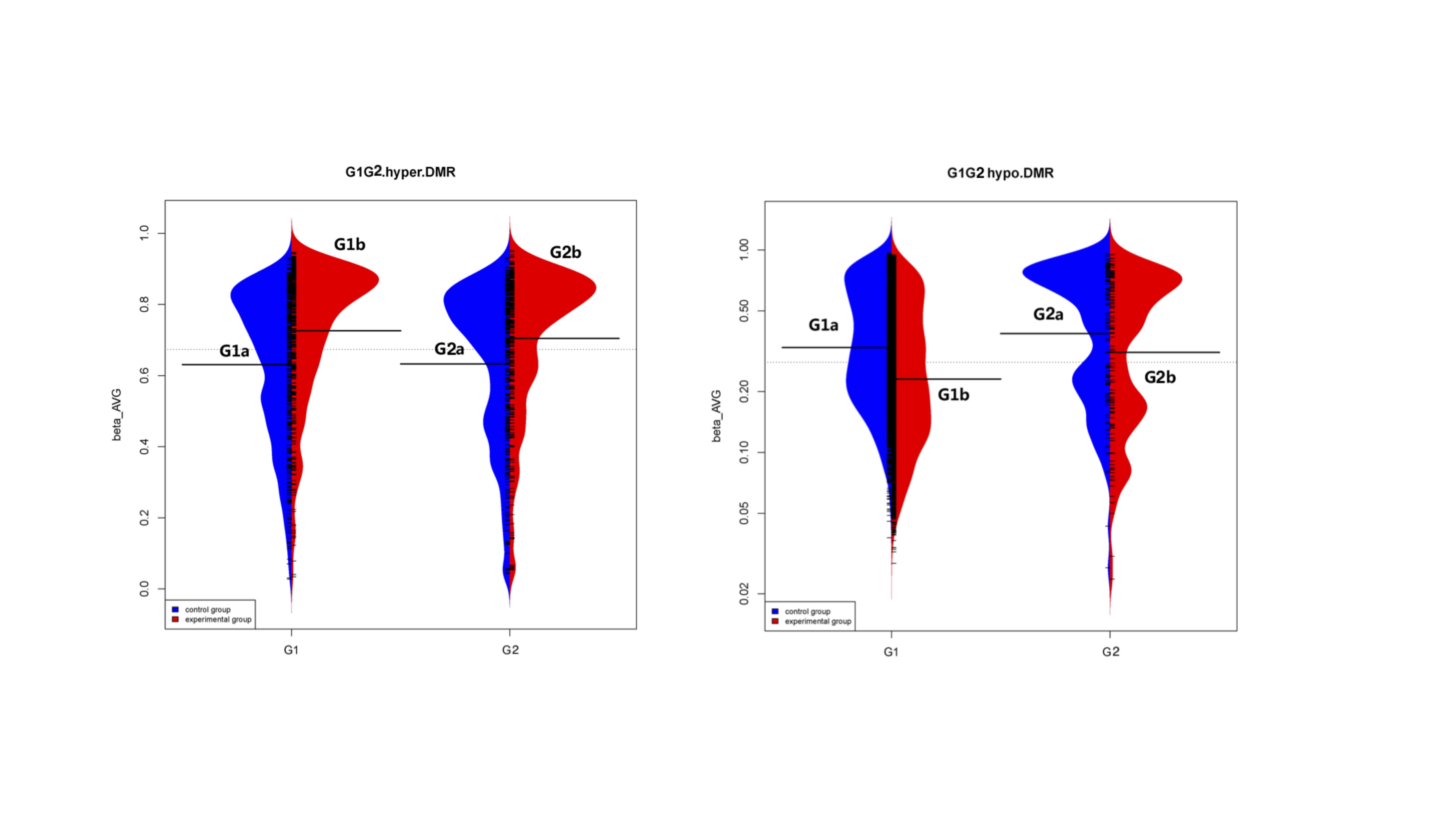
*Methods: The patients whose grafted kidney dysfunction by reason of acute rejection were enrolled as group 1(G1), where the mean survival time was around 2 years. The patients whose grafted kidney have survived for many years were recruited as group 2(G2). The PBMC of these patients before transplantation (G1a and G2a) and when their grafted kidney dysfunction/function at 2 years (G1b and G2b) respectively were sought out from the specimen bank. The matching healthy individuals were set as group 3(G3). The Illumina HumanMethylation450 BeadChip array was used to quantify the methylation status of each sample.
*Results: This study revealed the significant changes (approximately 60% of all CpG sites) in DNA methylation between ESRD (G1a and G2a) and health individuals (G3). These CpG sites mainly spread across TSS (21.72%), Body (33.97%), and IGR (22.14%) and mainly located on chromosome 1, 2 and 6, which was the same as the pattern of the comparison between G1a and G1b. Further KEGG pathways showing enrichment primarily included: Pathways in cancer and TGF-β signaling pathway. There was no obvious difference in the direct comparison of G1b and G2b. However, we found that the delta beta value of the overall mean methylation levels of hyper/hoper-methylated differentially methylated regions(DMRs) in G1a vs G1b was greater than that in G2a vs G2b. KEGG analysis showed an enrichment of these hyper/hypo-methylated DMRs related to pathways in cancer, HTLV-I infection, cell adhesion and calcium signaling pathway, which was inconsistent with that in G2a vs G2b.
*Conclusions: Our analysis revealed that there was a striking difference in the comparison of ESRD vs Health and the comparison before and after graft failure, which was mainly mapping to chromosome 1, 2 and 6. The targeted patients with graft failure manifested the markedly different pattern in the changed levels of DNA methylation and the enrichment of KEGG pathways. Hence, we suggested that DNA methylation may be another promising diagnostic and therapeutic method for patients with ESRD and grafted kidney.
To cite this abstract in AMA style:
Zhu C, Jiang H, Chen J. The Study on the DNA Methylation Dynamic Changes after Renal Transplantation [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/the-study-on-the-dna-methylation-dynamic-changes-after-renal-transplantation/. Accessed March 9, 2026.« Back to 2019 American Transplant Congress