The Complexity of T Cell-Mediated Rejection Scenarios: Identifying BK in Biopsies with TCMR
1University of Alberta, Edmonton AB, AB, Canada, 2Virginia Commonwealth University, Richmond, VA, 3Alberta Transplant Applied Genomics Centre, Edmonton, AB, Canada
Meeting: 2019 American Transplant Congress
Abstract number: 533
Keywords: Gene expression, Kidney, Polyma virus, Rejection
Session Information
Session Name: Concurrent Session: Kidney: Polyoma
Session Type: Concurrent Session
Date: Tuesday, June 4, 2019
Session Time: 4:30pm-6:00pm
 Presentation Time: 5:18pm-5:30pm
Presentation Time: 5:18pm-5:30pm
Location: Ballroom B
*Purpose: Some transplanted kidneys have concomitant T cell-mediated rejection (TCMR) and BK virus nephropathy (BK). The histologic diagnosis in BK sometimes overlooks the TCMR component, and molecular TCMR is often accompanied by BK, confusing interpretation. We examined molecular methods for identifying BK in biopsies with molecular TCMR in the INTERCOMEX study. (ClinialTrials.gov NCT01299168)
*Methods: Affymetrix microarray chips were used to build a molecular classifier to distinguish biopsies with BK virus from all others (N=1679, 55 with BK and 195 with histologic TCMR). The same analysis was repeated in all biopsies with molecular TCMR (N=192, 30 with BK). Molecular diagnoses were assigned with 10-fold cross-validation using the glmnet package in R. All steps, including probe set selection, were carried out from scratch within each cross-validation training set. These classifiers were used in conjunction with published classifiers for TCMR.
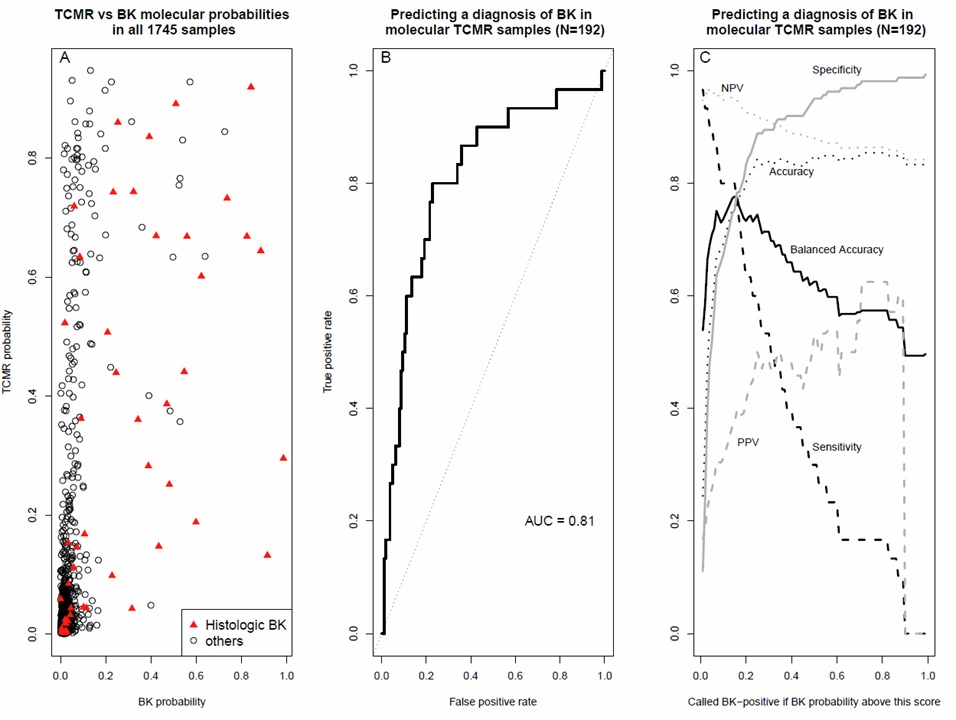
*Results: The top differentially expressed genes between TCMR and BK were higher in BK, and included PIF1 (5′-To-3′ DNA Helicase, which suppresses genome instability), CCDC74A (Coiled-Coil Domain Containing 74A), and immunoglobulin genes (IGHM and IGLL5) representing a plasma cell infiltrate. While most biopsies with high molecular BK scores had histologic BK, there was considerable overlap with molecular TCMR; some BK biopsies had low molecular BK scores (Figure 1A). The area under the curve (AUC) for BK vs all other diagnoses was 0.82 in the entire population (not shown) and 0.81 in the molecular TCMR population (Figure 1B). At the optimal cutoff (0.15 on the x-axis of Figure 1C), the balanced accuracy (mean of sensitivity and specificity) within the molecular TCMR population was ~0.77.
*Conclusions: The diagnostic accuracy of the classifier using human mRNAs on the current microarray is not sufficiently high to allow reliable separation between kidneys with and without BK (although measurement of viral mRNAs will probably resolve this – research in progress). However, given the relatively high AUC, the classifier can be used to flag biopsies with molecular TCMR that have a high probability of BK coexisting with TCMR.
To cite this abstract in AMA style:
Gupta G, Reeve J. The Complexity of T Cell-Mediated Rejection Scenarios: Identifying BK in Biopsies with TCMR [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/the-complexity-of-t-cell-mediated-rejection-scenarios-identifying-bk-in-biopsies-with-tcmr/. Accessed February 9, 2026.« Back to 2019 American Transplant Congress