Single Cell Analysis of Kidney Allograft Rejection
1Immunobiology, Cincinnati Children's Hospital, Cincinnati, OH, 2Biomedical Informatics, Cincinnati Children's Hospital, Cincinnati, OH, 3Internal Medicine, University of Cincinnati College of Medicine, Cincinnati, OH, 4Transplantation, University of Cincinnati College of Medicine, Cincinnati, OH
Meeting: 2019 American Transplant Congress
Abstract number: A4
Keywords: Gene expression, Genomics, Kidney, Rejection
Session Information
Session Name: Poster Session A: Acute Rejection
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: Rejection therapies (corticosteroids and antilymphocyte globulins) have not changed in over 50 years. In part this is due to a knowledge deficit of signaling pathways and cellular populations responsible for mediating allograft rejection. Thus, there is an urgent need to further understand the mechanisms underlying allograft rejection with a focus on potential therapeutic targets. While many studies have focused on analysis of peripheral blood, it is likely that allograft biopsy samples may yield more relevant information. Although bulk transcriptomic analysis of allograft biopsies has identified markers associated with acute rejection, bulk transcriptomics does not allow specific cellular identification of signaling pathways.
*Methods: Herein, we used novel freezing and tissue dissociation protocols and characterized renal allograft rejection using single cell RNA sequencing (scRNAseq). Using this approach, we extensively characterized allograft biopsies from normal living donor kidneys before transplant and several biopsies after late mixed rejection.
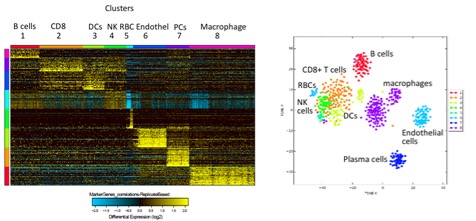
*Results: Notably, pre-transplant biopsies contained largely resident kidney cells (renal tubular epithelial and endothelial cells) and some resident immune cells (macrophages and a few T cells). During rejection we also observed resident kidney cells, but found a massive increase in infiltrating immune cells (CD4+, subsets of CD8+ T cells, B cells, and several subsets of macrophages). Interestingly, we identified a unique CD8+ T cell subpopulation that possesses low expression of granzyme B and perforin but high expression of granzyme K. To further restrict transcriptomic analysis to alloreactive T cells, we sequenced TCR α/β CDR3 regions in parallel with whole transcriptomes from the same cells. We identified several expanded clones within the allograft with a diverse range of gene expression.
*Conclusions: Pathway analysis of these expanded, presumably alloreactive T cell clones is underway and potential therapeutic targets will be tested in mixed lymphocyte reaction assays. Our approach can be tailored to each patient, correlated with potential biomarkers in the blood and urine, and provide a platform where therapeutic targets can be rationally defined, mechanistically-based and exploited.
To cite this abstract in AMA style:
Castro-Rojas CM, Roskin KM, Moore CG, Jawdeh BAbu, Salomonis N, Alloway RR, Woodle E, Hildeman DA. Single Cell Analysis of Kidney Allograft Rejection [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/single-cell-analysis-of-kidney-allograft-rejection/. Accessed February 19, 2026.« Back to 2019 American Transplant Congress