RNA Sequencing Identifies a Differential Role of Coding and Non-Coding Genes by Types of Rejection
1Department of Surgery, UCSF, San Francisco, CA, 2Department of Surgery, Bellvitge University Hospital, Barcelona, Spain, 3BCHSI, UCSF, San Francisco, CA
Meeting: 2019 American Transplant Congress
Abstract number: A175
Keywords: Gene expression, Kidney transplantation, Rejection
Session Information
Session Name: Poster Session A: Biomarkers, Immune Monitoring and Outcomes
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: The differentiation of histological phenotypes of T cell mediated cellular rejection (TCMR) and humoral or antibody mediated rejection (AMR) of kidney allografts, bears relevance on disparate clinical outcomes and different therapeutic approaches, and is important to better understand to improve injury reversal and mitigation of progression to untimely chronic immune-mediated transplant failure. Therefore, the study of the underlying molecular mechanisms of different pathological injuries has gained considerable interest to more specifically characterize the different pathological processes of rejection. Besides, the characterization of the molecular mechanisms of allograft rejection in peripheral blood as a non-invasive approach has also been a main endeavor among the transplant community. While most of the prior published transcriptional studies have allowed for a better understanding of the biology of kidney transplant rejection, these studies have been unable to identify distinct differences in molecular pathways regulating CMR and AMR.
*Methods: We harnessed RNA sequencing technology and applied advanced statistical techniques to characterize peripheral blood molecular pathways implicated during allograft rejection in kidney transplant patients displaying clear histological phenotypes of humoral (n=12), cellular (n=13) rejection or no injury/stable (n=12) kidney allograft paired biopsies.
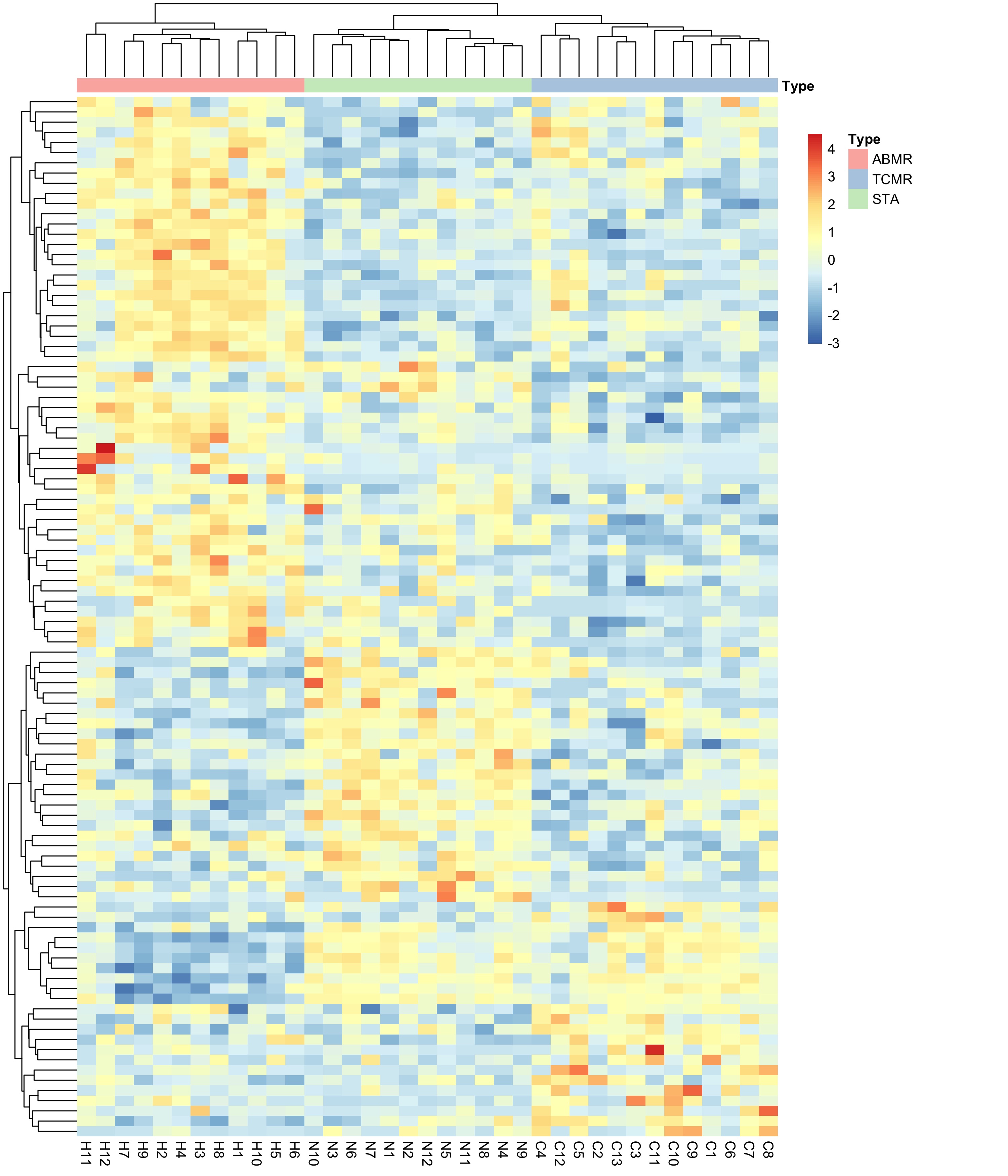
*Results: Applying ENET with a multinomial distribution to find differential expressed genes among the three clinical phenotypes, we found a set of 102 genes (62 protein-coding and 38 non-coding genes) (Figure 1) that perfectly separate the clinical phenotypes suggesting the different pathogenesis of kidney allograft rejection. Further exploration showed co-expression between coding and non-coding genes in AMR suggesting the role of non-coding RNA in the humoral rejection. In addition, we found AMR up-regulated genes significantly enriched (FDR<0.05) in several immunological signatures using gene set enrichment analysis. The top one is related with IRF4, which regulates Toll-like-receptor signaling.
*Conclusions: The abundance of non-coding RNA changes and its co-expression in AMR together with the immune signatures found suggests greater complexity of the immune response and its regulation in AMR. Overall, our findings highlight the complexity of the molecular mechanisms driving different histological and clinically separate categories of allograft rejection.
Figure 1. Heatmap showing the 102 genes associated with the three clinical phenotypes (AMR, TCMR and STA) by using ENET with a multinomial distribution.
To cite this abstract in AMA style:
Pineda S, Bestard O, Sigdel TK, Sirota M, Sarwal M. RNA Sequencing Identifies a Differential Role of Coding and Non-Coding Genes by Types of Rejection [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/rna-sequencing-identifies-a-differential-role-of-coding-and-non-coding-genes-by-types-of-rejection/. Accessed March 3, 2026.« Back to 2019 American Transplant Congress