RNA-Seq Based Analysis of Renal Allograft Biopsies Reveals Key Mediators of Interstitial Fibrosis/Tubular Atrophy
1Diabetes Complications Research Centre, Conway Institute, School of Medicine, University College Dublin, Dublin, Ireland
2North Dublin Renal Biobank, Dublin, Ireland
3Beaumont Hospital & RCSI, Dublin, Ireland
4University Health Network, Toronto, Canada.
Meeting: 2018 American Transplant Congress
Abstract number: 311
Keywords: Fibrosis, Gene expression, Kidney transplantation
Session Information
Session Name: Concurrent Session: Kidney Complications: Diagnostic Considerations
Session Type: Concurrent Session
Date: Monday, June 4, 2018
Session Time: 4:30pm-6:00pm
 Presentation Time: 4:54pm-5:06pm
Presentation Time: 4:54pm-5:06pm
Location: Room 6C
Interstitial Fibrosis/Tubular Atrophy(IFTA) is the final common pathway of most progressive renal diseases and is a potent predictors of renal outcome in both native and transplanted kidneys. IF/TA is a major cause of late allograft loss. Our understanding of the molecular mechanisms underpinning this complex process remains incomplete.
We performed RNA-Seq gene expression profiling on renal tissue obtained from transplanted patients undergoing a clinically indicated biopsy (n=21). IF/TA was determined from the renal biopsy report, and by undertaking quantitative morphometric analysis. We identified a subset of 671 genes differentially expressed (DE) (381 unregulated, 236 downregulated; FDR<0.1) between severe and mild/moderate IF/TA in our cohort, and used a systems biology approach to determine key mediators in this process.
Pathway Analysis reveals that B-Cell development, NRF2-mediated oxidative stress response & superoxide radical degredation comprise the top canonical pathways associated with the perturbed genes in our dataset. Pivotal upstream regulators are associated with lymphoid cell development (SPI1), proteasomal degradation (BACH1,LONP1) & regulation of antioxidant proteins (NFE2L2).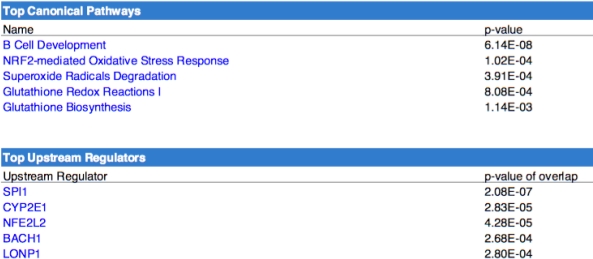 In silico cell-type enrichment analysis reveals enhanced transcriptional signatures from CD19+ B-Cells, Dendritic cells & CD4+ T-cells in the expression profiles of patients with severe IF/TA.
In silico cell-type enrichment analysis reveals enhanced transcriptional signatures from CD19+ B-Cells, Dendritic cells & CD4+ T-cells in the expression profiles of patients with severe IF/TA.
DE genes were used to generate a gene-protein interaction network for further analysis. Zero & first order interactions were examined. Significant gene modules detected in the networks are enriched for B-Cell signalling, ECM-Receptor interaction & chemokine signalling pathways.
These data underscore the prominent role of inflammation in the development of IF/TA and reveal novel potential mediators of this process.
CITATION INFORMATION: McEvoy C., Brennan E., Dorman A., Sadlier D., Konvalinka A., Conlon P., Godson C. RNA-Seq Based Analysis of Renal Allograft Biopsies Reveals Key Mediators of Interstitial Fibrosis/Tubular Atrophy Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
McEvoy C, Brennan E, Dorman A, Sadlier D, Konvalinka A, Conlon P, Godson C. RNA-Seq Based Analysis of Renal Allograft Biopsies Reveals Key Mediators of Interstitial Fibrosis/Tubular Atrophy [abstract]. https://atcmeetingabstracts.com/abstract/rna-seq-based-analysis-of-renal-allograft-biopsies-reveals-key-mediators-of-interstitial-fibrosis-tubular-atrophy/. Accessed February 25, 2026.« Back to 2018 American Transplant Congress
