RNA Expression Profiling of Renal Allografts in the Cynomolgus Monkey Identifies Tolerance
1Pathology, Massachusetts General Hospital, Boston, MA
2Laboratory Medicine and Pathology, University of Alberta, Edmonton, AB, Canada
3Department of Surgery, Massachusetts General Hospital, Boston, MA.
Meeting: 2018 American Transplant Congress
Abstract number: 526
Keywords: Bone marrow transplantation, Kidney transplantation, Tolerance
Session Information
Session Name: Concurrent Session: Kidney Immunosuppression: General Considerations - 2
Session Type: Concurrent Session
Date: Tuesday, June 5, 2018
Session Time: 4:30pm-6:00pm
 Presentation Time: 5:18pm-5:30pm
Presentation Time: 5:18pm-5:30pm
Location: Room 6A
Introduction
Tolerance induction is a long standing clinical goal. However, identification remains elusive. Here we profile intra-renal allograft RNA expression in a mixed chimerism renal allograft model in Cynomolgus monkeys and identify biologically significant tolerance.
Methods
Samples from 76 animals (one to 11 biopsies, zero to 5983 days post-transplantation. n = 278) included protocol, indication,plus autopsy nephrectomies. 21.4 % lacked a terminal rejection (tolerant), and 78.6 % showed terminal rejection. The gene set includes 67 oligonucleotides derived from human studies. JMP13 programs included Fit Model Platform, K-means clustering, Factor Analysis. P values were adjusted for false discovery. Factor analysis was chosen to reduce the many numerated gene expressions into smaller sets of correlated variables
Results
Factor analysis identified three dominant factors (Eigen >6), each with a different pattern of gene expression, relating to TCMR, CAMR, or Tolerance. The Tolerance factor comprises a novel set of gene expressions (Treg pathway by Gene Ontogeny). Clustering these three factors created nine groups. Calculated both per sample and per animal one of the nine clustered groups, the Tolerance cluster, showed the lowest probability of terminal rejection (P < 1-E-4), the longest duration of allograft survival (P < 1-E-3), and the lowest relative risk of terminal rejection (P < 1-E-3). The Tolerance factor could not be identified within current pathological diagnostic categories. The TCMR and CAMR factors are dominant to the Tolerance factor causing rejection even if the Tolerance factor is present. These three factors determine the probability of terminal rejection or tolerance. The figure shows the probability of survival for the clustered groups (A per sample, B per animal).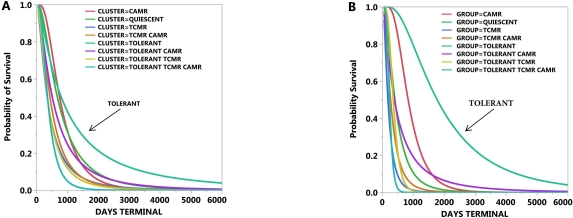 Conclusions
Conclusions
This novel a posteriori approach permits identification of pathways of rejection, including tolerance.
CITATION INFORMATION: Smith R., Matsunami M., Adam B., Rosales A., Oura T., Cosimi A., Kawai T., Mengel M., Colvin R. RNA Expression Profiling of Renal Allografts in the Cynomolgus Monkey Identifies Tolerance Am J Transplant. 2017;17 (suppl 3).
To cite this abstract in AMA style:
Smith R, Matsunami M, Adam B, Rosales A, Oura T, Cosimi A, Kawai T, Mengel M, Colvin R. RNA Expression Profiling of Renal Allografts in the Cynomolgus Monkey Identifies Tolerance [abstract]. https://atcmeetingabstracts.com/abstract/rna-expression-profiling-of-renal-allografts-in-the-cynomolgus-monkey-identifies-tolerance/. Accessed February 18, 2026.« Back to 2018 American Transplant Congress
