Rejection After Liver Transplantation: What Can We Learn from Transcriptomic Analysis?
1University of California, San Francisco, San Francisco, CA, 2UCLA, Los Angeles, CA
Meeting: 2022 American Transplant Congress
Abstract number: 579
Keywords: Gene expression, Rejection
Topic: Basic Science » Basic Clinical Science » 19 - Chronic Organ Rejection
Session Information
Session Name: Chronic Organ Rejection
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 7, 2022
Session Time: 5:30pm-7:00pm
 Presentation Time: 6:50pm-7:00pm
Presentation Time: 6:50pm-7:00pm
Location: Hynes Room 309
*Purpose: Acute cellular rejection (ACR) can occur at any time following liver transplantation (LT); initial ACR within 6 months is most common and occurs in up to 70% of recipients, with approximately 20% of patients experiencing one or more recurrent ACR episodes. Chronic rejection (CR) has been associated with increasing numbers of ACR episodes in some studies, and is the ultimate cause of graft failure in 15% of LT recipients. We sought to determine whether there were transcriptomic differences in ACR and CR after LT.
*Methods: A total of 38 formalin fixed paraffin embedded (FFPE) liver biopsy specimens were included (9 normal liver tissue, 21 ACR, 8 CR). Specimens from recipients with HBV, HCV, or autoimmune hepatitis were excluded. RNA was isolated from FFPE sections, and gene expression was measured using the NanoString platform. Differential gene expression (DE) analysis and pathway analysis (PA) using the KEGG database were performed, with Benjamini Hochberg p-value correction for false discovery rate.
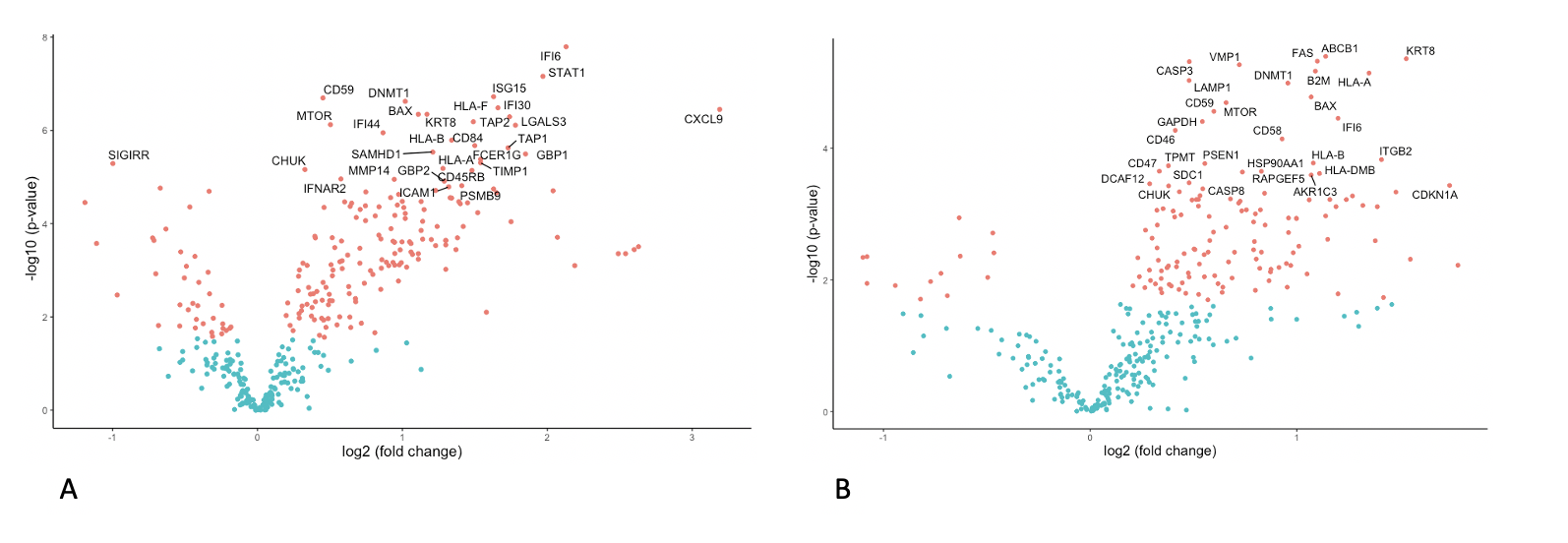
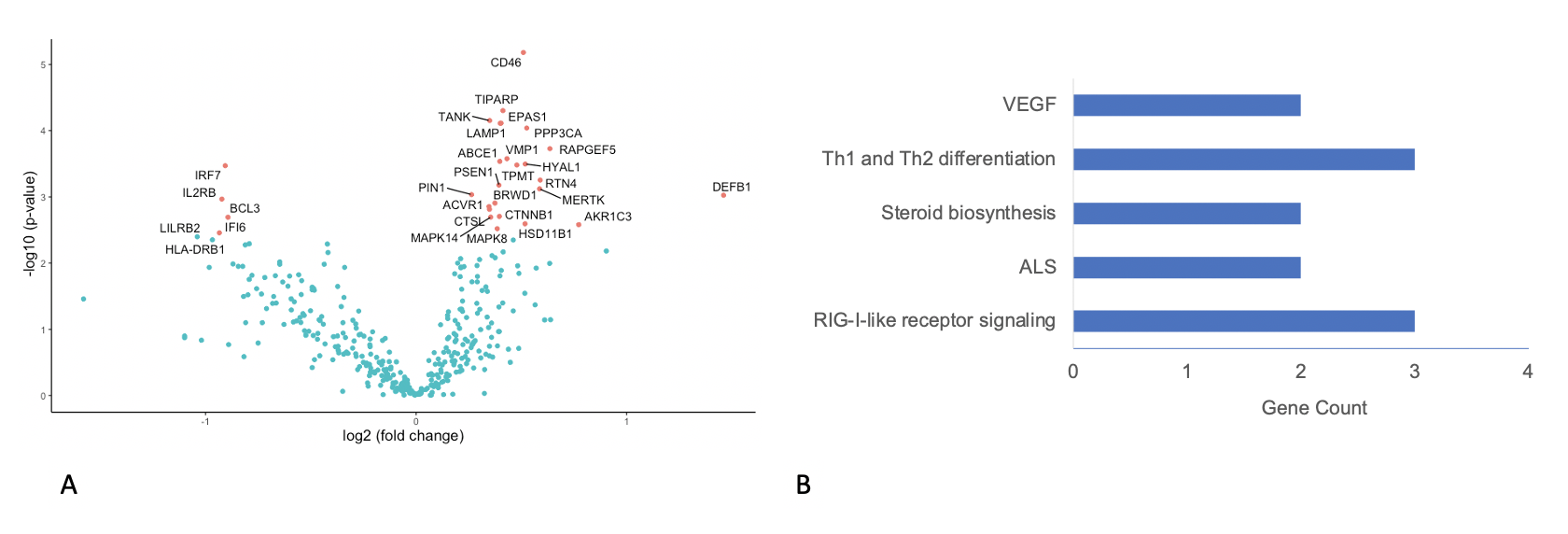
*Results: There were significant differences in DE between ACR and normal liver tissue (Figure 1A, 142 genes), and in PA there was significant upregulation of genes involved in the common rejection module (n=4 genes) and antigen processing and presentation in ACR versus normal (n=15 genes, p<0.001). Similarly, there were significant differences in DE between CR and normal (Figure 1B, 145 genes) and PA showed significant upregulation of genes involved in antigen processing and presentation (n=8) and allograft rejection (n=6 genes). In DE comparing ACR and CR, significant differences in 28 genes were identified (Figure 2A), with PA demonstrating small differences in a variety of immunologic pathways (Figure 2B).
*Conclusions: Compared with normal liver allograft biopsies, both acute and chronic rejection show significant enrichment of pathways associated with allograft rejection and antigen processing. Only acute rejection showed overexpression of genes present in the common rejection module. Our hope is that these findings will serve as the foundation for identifying unique pathways associated with chronic rejection leading to better or novel strategies for immunotherapy.
To cite this abstract in AMA style:
Braun HJ, Amara D, Zarinsefat A, Szabo G, Laszik Z, Stock P, Ascher N. Rejection After Liver Transplantation: What Can We Learn from Transcriptomic Analysis? [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/rejection-after-liver-transplantation-what-can-we-learn-from-transcriptomic-analysis/. Accessed February 21, 2026.« Back to 2022 American Transplant Congress