Quantitative Proteome Profiling in T Cell-Mediated Rejection
1Department of Urological Organ Transplantation, The Second Xiangya Hospital, Central-South University, Changsha, China, 2Department of Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, 3Department of Biostatistics, Graduate School of Public Health, and Departments of Pharmacology and P, University of Pittsburgh Medical Center, Pittsburgh, PA, 4Department of Pharmacology and Chemical Biology, School of Medicine, University of Pittsburgh, Pittsburgh, PA
Meeting: 2019 American Transplant Congress
Abstract number: A8
Keywords: Biopsy, Kidney transplantation, Rejection, T cells
Session Information
Session Name: Poster Session A: Acute Rejection
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: The proteomic analyses through formalin-fixation and paraffin embedding (FFPE) specimens reflects systemic and inherent renal injury perturbations for different kidney transplant injury states. In this study, we develop a high-sensitivity and high-confidence workflow to map and decipher proteome changes from the limited amount of FFPE samples after renal transplantation.
*Methods: 5 patients with T cell-mediated rejection (TCMR), and 5 normal healthy controls (HC). Quantitative analysis of the FFPE proteome was performed using loss-less basic pH reverse phase fractionation in combination with TMT based mass spectrometry (MS) (n=3). With the application of quantile normalization followed by limma statistic test for the differential expression analysis, potential biomarkers with high confidence were screened out. Based on the peptidome of the biomarker candidates, MS based selected reaction monitoring method (SRM) for validation and machine learning approach.
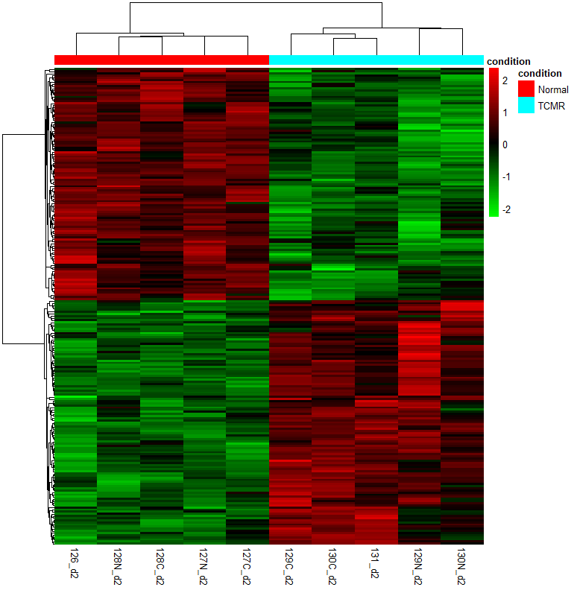
*Results: Based on our high-sensitivity sample preparation workflow and MS instrument, we were able to successfully detect and quantify 2498 proteins from TCMR samples. Comparison of TCMR and normal samples uncovered 133 differential expression (DE) proteins (FDR<0.01) in all three replicated datasets. 77 were only present in TCMR group. Among them, 33 up-regulation DE proteins and 41 down-regulation DE proteins were taken into consideration with a strict statistic criteria. Further analysis revealed that 137 biological process, 42 cell components, 106 molecular functions and 47 pathways in the down-regulation DE protein group together with 865 biological process, 183 cell components, 74 molecular functions and 100 pathways in the up-regulation DE protein group were respectively dig out to clarify the whole pathophysiological procedures of TCMR. Interestingly, PD-1 signaling and phosphorylation of CD3 and TCR pathways which are participated in antigen presentation are close related with the up-regulation of PDIA3 and PTPRC, the downstream functions of both proteins are dependent on HLA-DPB1.
*Conclusions: A high-sensitivity and high-confident clinical proteomics workflow was developed to analyze a large fraction of the disease-related proteome from small amounts of FFPE samples. This proteome research of biomarker discovery and validation with mass spectrometry can identify protein fingerprints for different kidney transplant injuries, based on which a robust assay will be developed to open the door for personalized immune risk assessment and therapy.
To cite this abstract in AMA style:
Song L, Zeng G, Liu P, Fang F, Liu H, Xie X, Randhawa P, Xiao K. Quantitative Proteome Profiling in T Cell-Mediated Rejection [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/quantitative-proteome-profiling-in-t-cell-mediated-rejection/. Accessed February 18, 2026.« Back to 2019 American Transplant Congress