Precision Medicine in Renal Transplantation: Structural Biology of HLA Defines Allelic Binding of Cardinal Pathogenic Viruses in Transplantation
K. R. Sherwood1, A. Nguyen2, J. Tran1, O. Gunther3, A. Nellore2, R. Thompson2, J. Lan1, L. Allan1, P. A. Keown1
1Vancouver General Hospital, Vancouver, BC, Canada, 2Biomedical Engineering, Oregon Health & Science University, Portland, OR, 3Gunther Analytics, Vancouver, BC, Canada
Meeting: 2021 American Transplant Congress
Abstract number: LB 16
Keywords: Antigen presentation, Histocompatibility antigens, Immunogenicity
Topic: Clinical Science » Infectious Disease » All Infections (Excluding Kidney & Viral Hepatitis)
Session Information
Session Name: Late Breaking: Basic & ID
Session Type: Rapid Fire Oral Abstract
Date: Monday, June 7, 2021
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:35pm-6:40pm
Presentation Time: 6:35pm-6:40pm
Location: Virtual
*Purpose: The incidence, clearance and clinical expression of viremia post-Tx are heterogeneous, complicating prediction, prognosis and therapy. We postulated that variation viral presentation related to HLA type may inform understanding, and we used in silico methods to assess the MHC-peptide binding affinity for three important phylogenetically distinct viruses (SARS-CoV-2, CMV and BKV).
*Methods: Carrier frequencies for 11 HLA genes (HLA-A, B, C; DRB1, DRB3/4/5, DQA1, DQB1, DPA1, DPB1) were determined by NGS in 1150 renal transplant recipients. All FASTA-formatted viral protein sequence data from the NCBI RefSeq database were kmerized into 8-12 mers. Using netMHCpan, MHC-peptide binding affinities were predicted and affinity scores <500nM were included in the HLA allele rankings
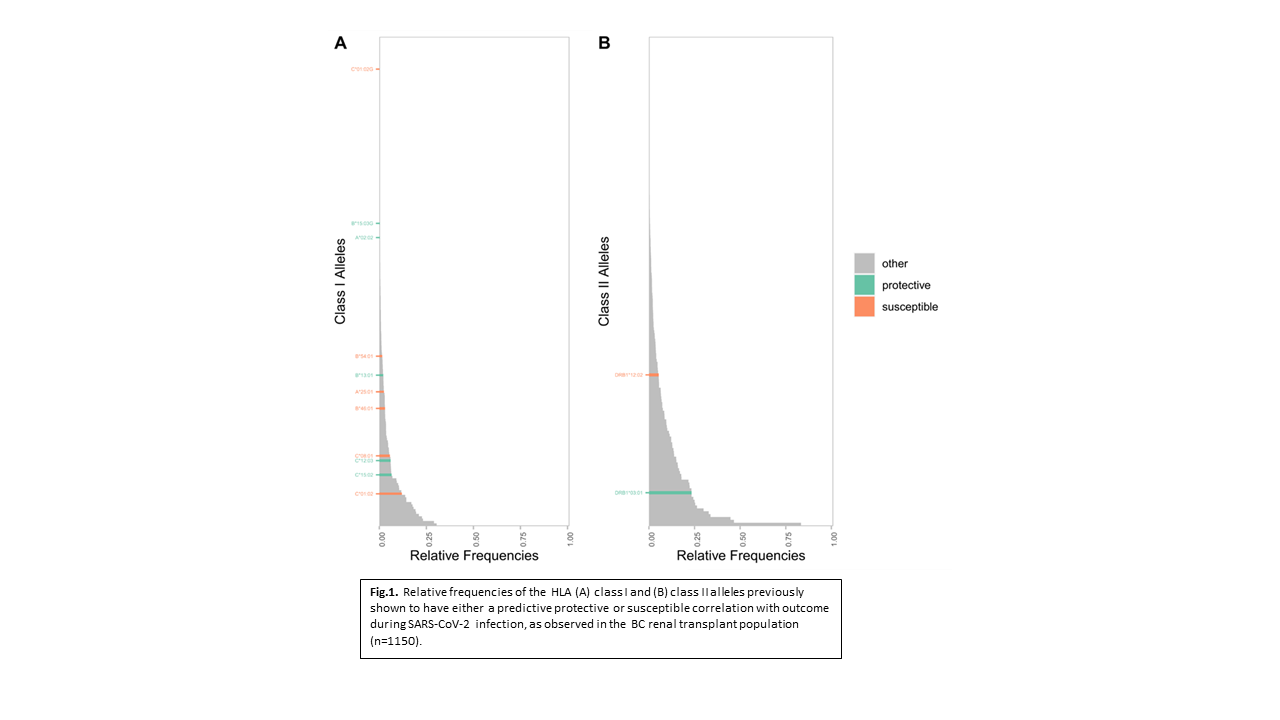
*Results: A total of 206 Class I HLA alleles identified in 1150 patients exhibited population frequencies ranging from 0.09% to 30%. Within this repertoire, peptide binding varied dramatically identifying low- and high-affinity alleles for each of the three viral proteomes. Alleles with lowest binding propensity were specific to each virus (e.g. HLA-B*46:01 for SARS-CoV-2, HLA-B*51:05 for CMV and HLA-B*15:08 for BKV). In contrast, alleles with the highest binding propensity were remarkably uniform for all 3 viruses (e.g. HLA-A*02:11 top for all three virus proteomes). Sequence signatures for HLA isoforms in the same allele group defined high or low binding characteristics, the difference being conferred by as little as a single amino acid within the peptide binding region (e.g. HLA-B*15:08 vs B*15:03). Carrier rates for predicted SARS-CoV-2 susceptibility/resistance genes in the transplant population were observed to be similar to global population carrier rates (Fig 1).
*Conclusions: Evaluation of peptide binding provides a unique insight into viral recognition. If confirmed in our current proof-of-principle study comparing sequence and outcome in a large Canadian population, this data may offer a vital biomarker to define risk and treatment within conventional virological patient strata following transplant.
To cite this abstract in AMA style:
Sherwood KR, Nguyen A, Tran J, Gunther O, Nellore A, Thompson R, Lan J, Allan L, Keown PA. Precision Medicine in Renal Transplantation: Structural Biology of HLA Defines Allelic Binding of Cardinal Pathogenic Viruses in Transplantation [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/precision-medicine-in-renal-transplantation-structural-biology-of-hla-defines-allelic-binding-of-cardinal-pathogenic-viruses-in-transplantation/. Accessed February 21, 2026.« Back to 2021 American Transplant Congress