One Nuclei at a Time: Decoding Kidney Allograft Fibrosis by Single Nuclei RNA-Sequencing
1University of Maryland, Baltimore, Baltimore, MD, 2University of Tennessee Health Science Center, Memphis, TN, 3University of Maryland, Baltimore, MD, 4UTHSC, memphis, TN, 5University of Tennessee/Methodist Transplant Institute, Memphis, TN, 6Weill Cornell Medicine, New York, NY
Meeting: 2022 American Transplant Congress
Abstract number: 575
Keywords: Fibrosis, Kidney, Kidney transplantation
Topic: Basic Science » Basic Clinical Science » 19 - Chronic Organ Rejection
Session Information
Session Name: Chronic Organ Rejection
Session Type: Rapid Fire Oral Abstract
Date: Tuesday, June 7, 2022
Session Time: 5:30pm-7:00pm
 Presentation Time: 6:10pm-6:20pm
Presentation Time: 6:10pm-6:20pm
Location: Hynes Room 309
*Purpose: The biological mechanisms underpinning late graft loss due to chronic allograft dysfunction remain unknown. Single nuclei RNA-sequencing (snRNAseq) of healthy and fibrotic kidney grafts has the potential to provide high resolution of cellular identities as well as a better understanding of individual cells function in the context of fibrosis.
*Methods: SnRNAseq was used to study histologically classified normal (N=3) and IFTA (N=5) human allografts. All allografts included in this study were functional and ≥15mos posttransplant. Analysis of each dataset was generated with the CellRanger 4.0 software. Dimensionality reduction was applied and visualized using UMAP by the Seurat pipeline. DEGs (FDR≤0.05 and FC≥1.5) identified significantly enriched GO terms and pathways. Trajectory analysis was performed using Monocle 3.
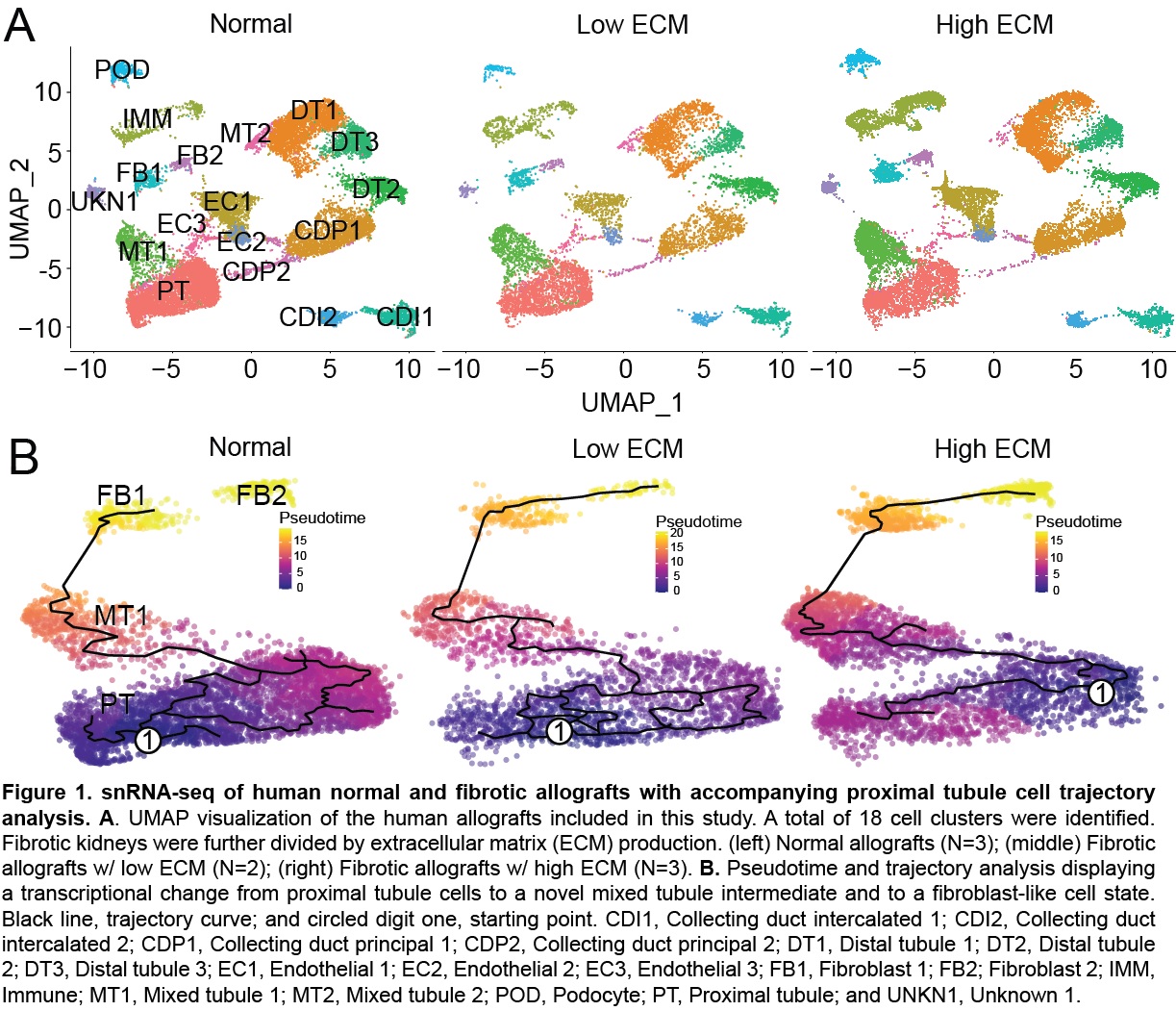
*Results: A total of 41,893 cells from 8 human kidney biopsies were sequenced. Transcriptomic analysis identified two forms of fibrosis with varying degrees of extracellular matrix (ECM) production, high and low ECM (Fig 1A). High ECM co-expressed high levels of CUBN, HAVCR1, MKI67, exhibiting injury-induced replicative repair of proximal tubules, whereas low ECM co-expressed high levels of HAVCR1 and low levels of MKI67, suggesting maladaptive repair. Low ECM exhibited greater G2M arrest and metabolic dysfunction. Trajectory analysis revealed a novel cell state, mixed tubule 1 (MT1) or injured proximal tubules (Fig 1B). Fibrotic-derived epithelial cells undergoing epithelial mesenchymal transition passed through the MT1 intermediate to a transcriptionally active fibroblast state. The endothelial mesenchymal (endoMT) transition was observed through the expression of partial endoMT markers, ZEB1/2 and PECAM, and fibroblast markers, FN1 and VIM. GO analysis highlighted fatty acid metabolism and Wnt signaling as vital pathways of fibrosis.
*Conclusions: This is the first study to use human kidney allografts ≥15mos posttransplant to describe the cellular landscape of the fibrotic kidney. ECM deposition contributes to injury severity and, potentially, can be used to differentiate between regenerative or maladaptive epithelial repair. Such findings will contribute to the development of cell-specific treatments of kidney fibrosis.
To cite this abstract in AMA style:
McDaniels J, Shetty A, Kuscu C, Kuscu C, Bhardi E, Rousselle T, Drachenberg C, Talwar M, Eason J, Muthukumar T, Maluf D, Mas V. One Nuclei at a Time: Decoding Kidney Allograft Fibrosis by Single Nuclei RNA-Sequencing [abstract]. Am J Transplant. 2022; 22 (suppl 3). https://atcmeetingabstracts.com/abstract/one-nuclei-at-a-time-decoding-kidney-allograft-fibrosis-by-single-nuclei-rna-sequencing/. Accessed February 27, 2026.« Back to 2022 American Transplant Congress