Microarray and RNA-Seq Analysis of Rejection in Formalin-Fixed Paraffin-Embedded (FFPE) Biopsies: Comparing FFPE to Conventionally Stabilized (RNAlater Solution) Biopsies
1University of Alberta, Edmonton, AB, Canada, 2R&D Transplant Diagnostics Division, Thermo Fisher Scientific, Los Angeles, CA
Meeting: 2020 American Transplant Congress
Abstract number: A-323
Keywords: Gene expression, Kidney transplantation, Methodology
Session Information
Session Name: Poster Session A: Biomarker Discovery and Immune Modulation
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Molecular understanding of organ transplant biopsies requires genome-wide measurements and molecular classifiers. To assess potential of performing molecular diagnostics on formalin-fixed paraffin-embedded (FFPE) biopsies, we first compared performance of microarrays and RNA-Seq on biopsies stabilized in RNAlater solution, and then compared molecular phenotyping of kidney transplant rejection both by microarrays and RNA-Seq in biopsies preserved at the same time in RNAlater solution or in FFPE.
*Methods: To compare material stabilized in RNAlater solution by microarrays and RNA-Seq, 10 biopsies were selected to represent T cell-mediated and antibody-mediated rejection (TCMR, ABMR) vs. no rejection. We then selected 70 FFPE vs. RNAlater biopsies (from 68 patients) representing TCMR, ABMR and no rejection, and compared the microarray and RNA-Seq results, including newly generated molecular classifiers for TCMR and ABMR.
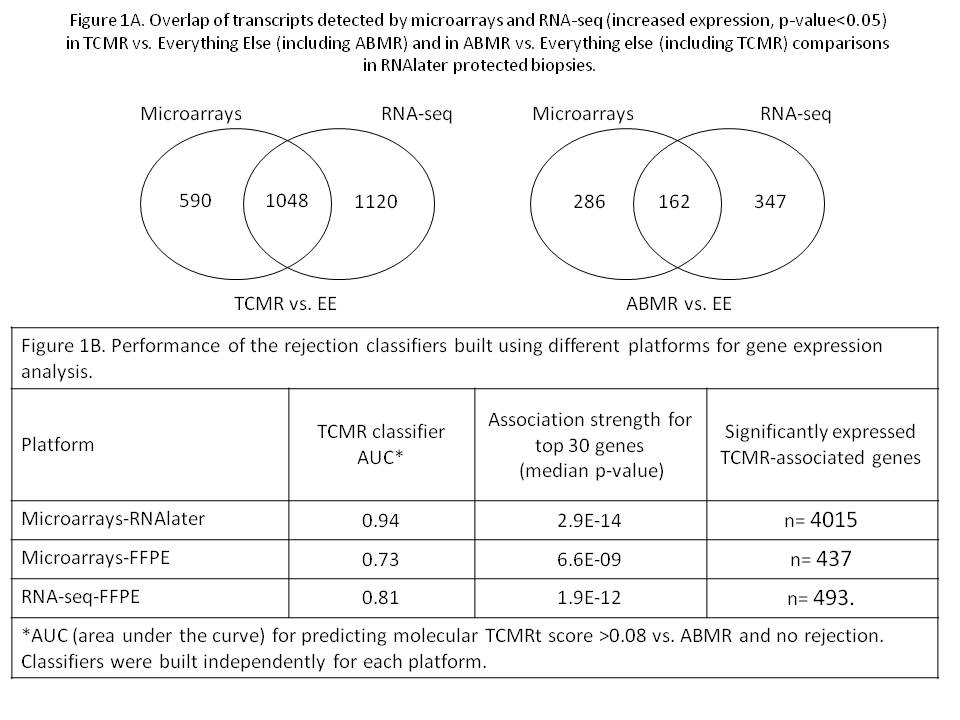
*Results: RNAseq and microarray results in RNAlater biopsies were similar, with substantial overlap of transcripts detected in TCMRvs.EE comparison (64%), with 80% overlap between the top 30 transcripts (Figure 1A). In ABMRvs.EE comparison the overlap was less (36%), with 73% overlap between the top 30 transcripts. RNA-Seq detected more rejection-associated transcripts than microarrays. Comparing FFPE to RNAlater biopsies (Fig. 1B), the strength of associations for rejection-associated transcripts was much less for FFPE either by microarrays or RNAseq than by microarray-RNAlater for TCMR and ABMR (not shown). AUCs for classifiers predicting TCMR were also better for microarray-RNAlater than in FFPE tissue (Figure 1B). Ten times fewer genes were significantly associated with TCMR in FFPE tissue at p-value<0.05. FFPE also induced artificial associations: a surprising overrepresentation of histone genes (5 of top 30 transcripts associated with TCMR by RNA-Seq-FFPE, indicating that FFPE alters the hierarchy of molecular associations. It is possible that histone transcripts are less sensitive to FFPE damage and therefore relatively overrepresented.
*Conclusions: Optimal development of molecular assessment should be based on tissue stabilized in RNAlater. FFPE destroys many transcripts and alters the hierarchy of molecular associations to create artificial associations, and RNA-Seq cannot circumvent these problems.
To cite this abstract in AMA style:
Halloran P, Famulski K, Reeve J, Huang J, Alexandrov N. Microarray and RNA-Seq Analysis of Rejection in Formalin-Fixed Paraffin-Embedded (FFPE) Biopsies: Comparing FFPE to Conventionally Stabilized (RNAlater Solution) Biopsies [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/microarray-and-rna-seq-analysis-of-rejection-in-formalin-fixed-paraffin-embedded-ffpe-biopsies-comparing-ffpe-to-conventionally-stabilized-rnalater-solution-biopsies/. Accessed March 9, 2026.« Back to 2020 American Transplant Congress