Machine Learning to Predict Deceased Donor Kidney Biopsy Results
United Network for Organ Sharing, Richmond, VA
Meeting: 2020 American Transplant Congress
Abstract number: B-013
Keywords: Area-under-curve (AUC), Biopsy, Kidney, Prediction models
Session Information
Session Name: Poster Session B: Kidney Deceased Donor Allocation
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Despite inconclusive evidence that kidney biopsy findings are associated with graft outcomes, biopsies are often cited as a key factor on whether to accept a deceased kidney offer. Motivated by the influential role of biopsy findings and to determine if biopsies are representations of other clinical data, we developed and evaluated a machine learning model to predict biopsy results given only clinical and lab data available from the OPTN at the time of match.
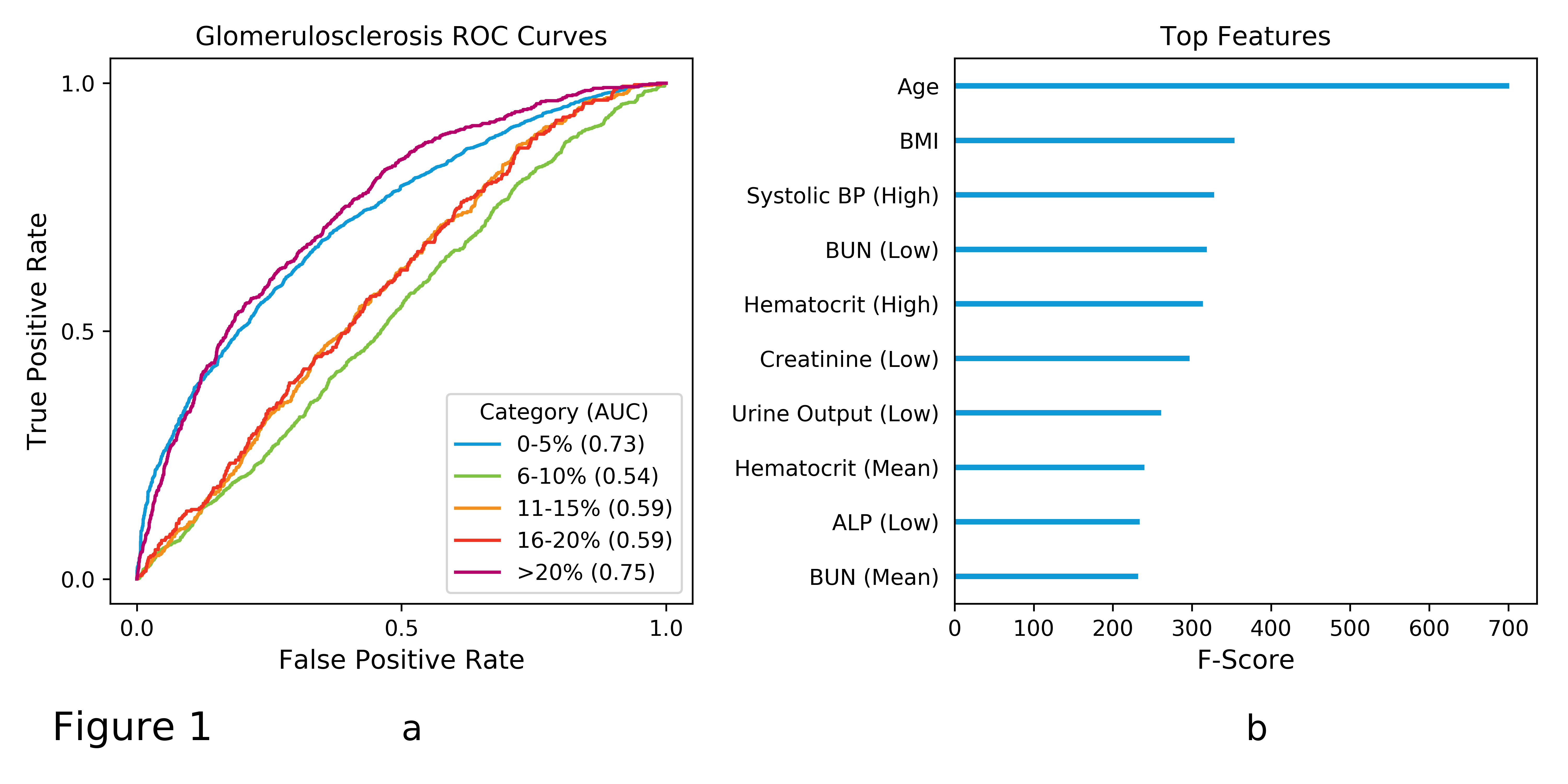
*Methods: We collected OPTN data, at the time of match, for deceased donors since 1999 with a kidney glomerulosclerosis (GS) (N = 63819 kidneys from 32968 donors) assessment. Next, we identified variables that could be associated with a kidney biopsy. Our dataset included mixed continuous/discrete demographic variables (e.g. age and ethnicity), discrete medical history indicators (e.g. history of hypertension), and continuous lab measurements (e.g. serum creatinine levels). After we generated the dataset, we trained an XGBoost machine learning model to predict the following GS categories: 0-5%, 6-10%, 11-15%, 15-20% or >20%. The model was trained on 90% of the data; the remaining 10% were reserved for model assessment via ROC analysis. We calculated ROC curves, Figure 1a, in a one-versus-rest manner (e.g. 20%+ GS versus all other categories).
*Results: We observed markedly different performance across categories. The GS model performs relatively well at rank-ordering on the extreme ends (0-5% and >20%; AUC 0.73 and 0.75) but poorly for intermediate categories (AUCs from 0.54 to 0.59). Among the top features, as determined from XGBoost feature importances, were age and BMI followed by a number of features derived from lab measurements (Figure 1b).
*Conclusions: Distinct differences in performance across GS categories may indicate challenges for automated biopsy prediction models. A key challenge is that the reliability between pathologists may vary depending on the level of GS. Hence, there may be lack of agreement from a single biopsy; however, due to a phenomenon known as restricted variance, one could expect better agreement at the extremes values and increased uncertainty toward the middle as observed in our model. Further investigation is needed to assess confidence of biopsy results stratified by severity and the impact on predictive models derived from those results.
To cite this abstract in AMA style:
Martinez C, Placona A. Machine Learning to Predict Deceased Donor Kidney Biopsy Results [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/machine-learning-to-predict-deceased-donor-kidney-biopsy-results/. Accessed February 22, 2026.« Back to 2020 American Transplant Congress