Irf8-Relevant Mechanism in B Cell Regulation In Vitro
1Organ Transplantation Center, and School of Medicine, University of Electronic Sciences and Technology of China, Chengdu, China, 2Center for Transplantation Sciences, MGH, Boston, MA
Meeting: 2020 American Transplant Congress
Abstract number: D-340
Keywords: B cells
Session Information
Session Name: Poster Session D: B-cell / Antibody /Autoimmunity
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Previously, we had reported that IFN regulatory factor 8 (IRF8) could be used for the identification of interstitial fibrosis and tubular atrophy (IFTA) with inflammation (IFTA-I) and could potentially serve as a prognostic marker for renal graft failure. Others have reported that IRF8 plays a crucial role in the normal development of marginal zone and follicular B cells. As an IRF8-relevant mechanism in IFTA-I is not yet clear, this study aims to identify potential regulatory factors and pathways in B cell development related to IRF8 by analyzing RNA-Seq data.
*Methods: Publicly shared transcriptional data of marginal zone B cells (MZB) and follicular B cells (FOB) from IRF8 conventional knockout (IRF8-/-) and Activation Induced Cytidine Deaminase knock out (AICDA-/-) mice were obtained from the Gene Expression Omnibus (GEO, GSE: 24972, and 47703). Analysis was performed using R programming packages (Oligo packages in R version: 3.6.1). Genes with Log2 |FC| > 1 and P < 0.05 were defined as the differentially expressed genes (DEGs). Targets of IRF8 was identified by ChIP-chip.
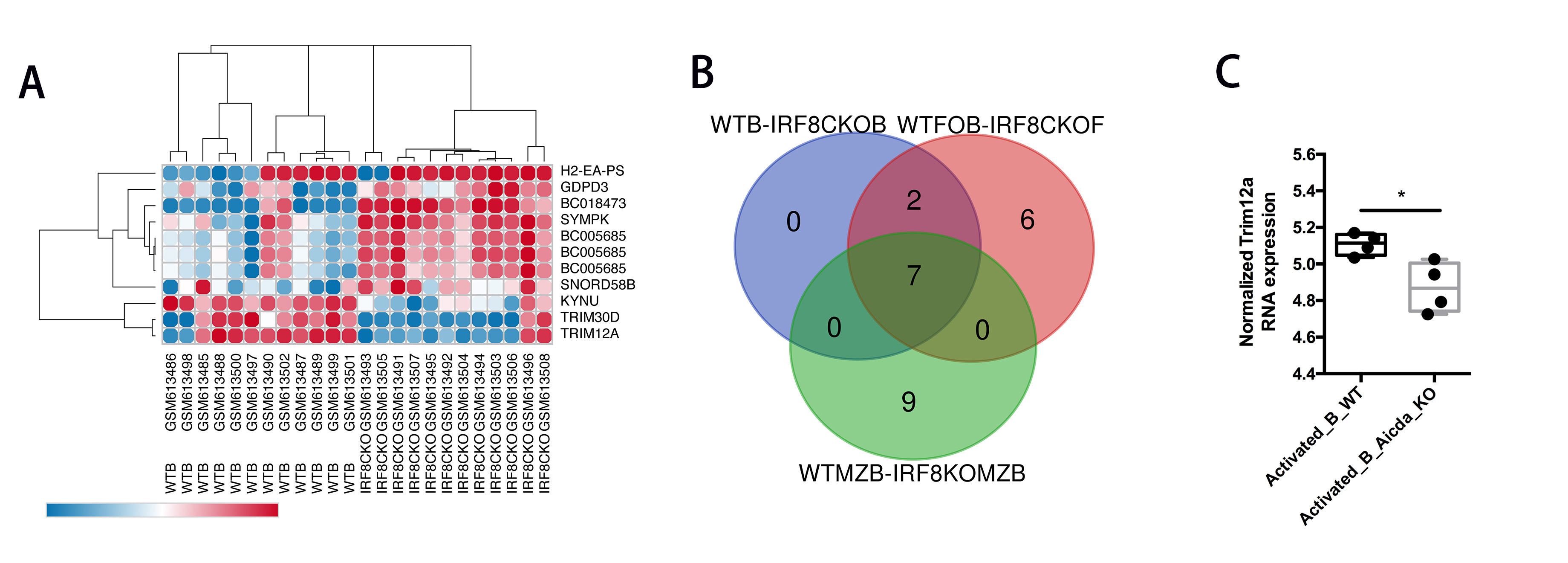
*Results: In comparison to a control group of WT mouse B cells (n=12), R analysis found 10 DEGs in the IRF8-/- B cell group (n = 12), among which 7 transcripts were up-regulated and 3 transcripts were down-regulated (Fig. A). Pearson correlation analysis found the transcriptional changes between WT B cells and IRF8-/- B cells to be significantly different. In comparison with the WT MZB cell group (n=6), there were 12 up-regulated transcripts and 4 down-regulated transcripts in the IRF8-/- MZB group (n = 6). In a similar comparison with the WT FOB cell group, there were 7 up-regulated and 8 down-regulated transcripts in the IRF8-/- FOB group. Among these three comparisons, 7 common DEGs were found (Fig. B), among which Trim12a was identified as a target of IRF8. Additionally, Trim12a expression was found to be significantly lower in AICDA knockout mice (Fig. C).
*Conclusions: Our study finds that Trim12a expression is related to the expression of IRF8 and AICDA genes. IRF8 may regulate AICDA expression through a Trim12a-relevant pathway that is found in the development of MZ and FO B cells and may play a role in renal graft failure. These results open room for further mechanistic studies regarding the role of IRF8 and Trim12a in transplantation immunology.
To cite this abstract in AMA style:
Fu Q, Deng K, Yang H, Markmann JF, Deng S. Irf8-Relevant Mechanism in B Cell Regulation In Vitro [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/irf8-relevant-mechanism-in-b-cell-regulation-in-vitro/. Accessed February 19, 2026.« Back to 2020 American Transplant Congress