Integrative Analysis of Immune Repertoire and Gene Expression from RNA-seq in Kidney Organ Transplants
1Department of Surgery, UCSF, San Francisco, CA, 2BCHSI, UCSF, San Francisco, CA, 3Department of Surgery, 3Bellvitge University Hospital, Barcelona, Spain
Meeting: 2019 American Transplant Congress
Abstract number: A5
Keywords: Gene expression, Kidney transplantation, Rejection, T cell receptors (TcR)
Session Information
Session Name: Poster Session A: Acute Rejection
Session Type: Poster Session
Date: Saturday, June 1, 2019
Session Time: 5:30pm-7:30pm
 Presentation Time: 5:30pm-7:30pm
Presentation Time: 5:30pm-7:30pm
Location: Hall C & D
*Purpose: The main problem with kidney transplantation is the possibility of rejection, which is classified as either antibody-mediated rejection (AMR) or T-cell-mediated rejection (TCMR) depending on the driving mechanism in the immune system. An organism’s immune system is determined by a set of B-cell receptor (BCR) and T-cell receptors (TCR) that are collectively known as the immune repertoire. Studying the role of the immune cells in rejection and its relationship with gene expression, new knowledge on the mechanism of rejection may be discovered. Immune repertoire has been conducted through the direct sequencing of the V(D)J recombination and complementary-determining region 3 (CDR3), however, with the advent of new computational approaches, adequate coverage of the immune repertoire can be extracted from RNA sequencing (RNAseq) data.
*Methods: We leverage RNAseq from peripheral blood of 12 AMR and 13 TCMR biopsy-confirmed rejections and 12 stables (STA) patients. We obtained BCR (heavy and light chain) and TCR (α, β, γ, δ) types and examine overall diversity, clonality and other types of features of the immune repertoire such as the α, β/γ, δ ratio and its association with the three clinical phenotypes. Then, we integrate these immune features with gene expression.
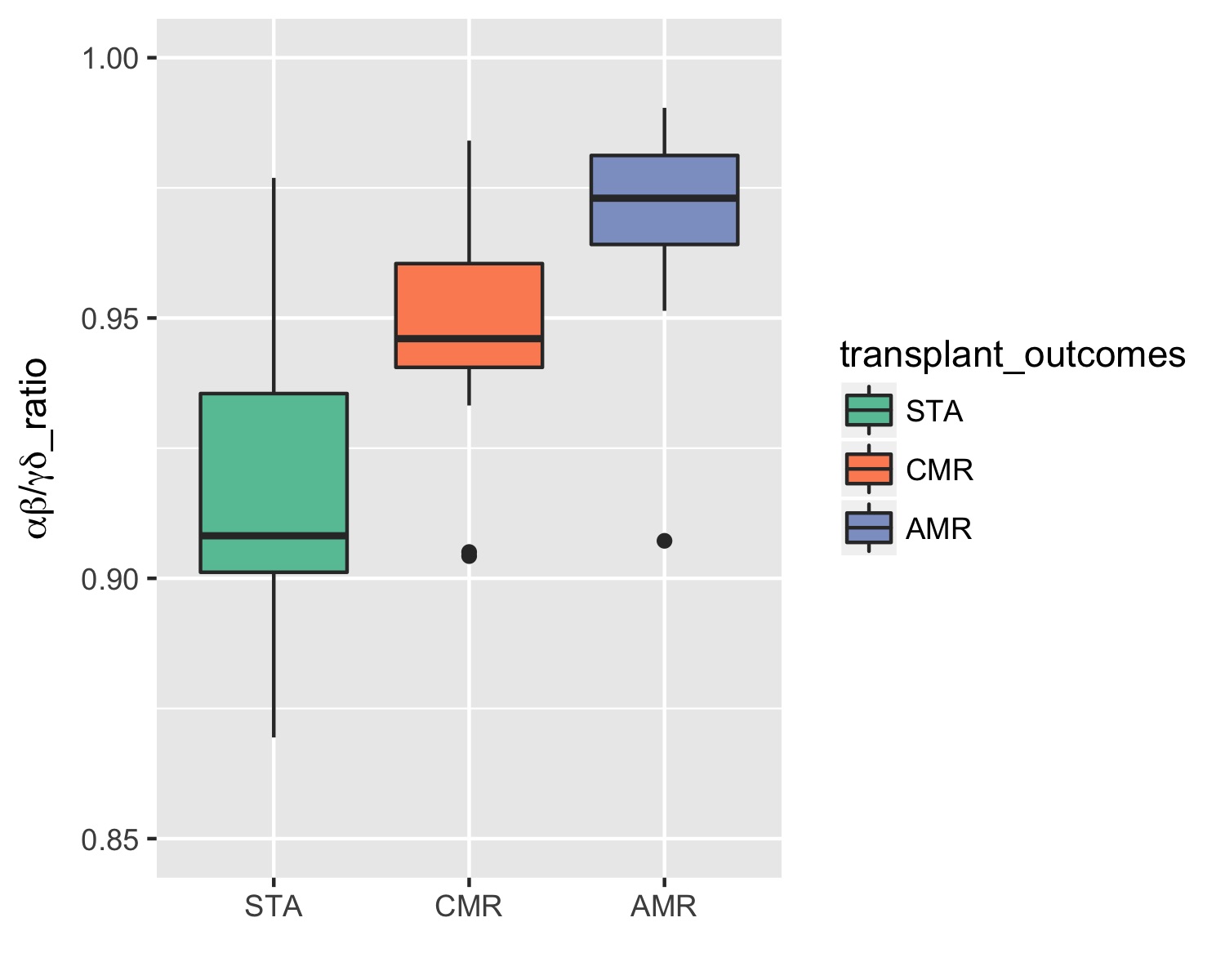
*Results: Among all the immune features, the α, β/γ, δ ratio was the most significant one, especially when comparing STA with AMR samples (p-value = 1.0*10-4). This ratio appears to be higher in transplant rejection samples (Figure 1) and was associated with 81 genes (25 downregulated and 56 upregulated) that were significantly enriched in allograft rejection pathway (p-value = 1.5*10-7), antigen processing and presentation (p-value = 1.2*10-5) and graft-vs-host disease (p-value = 2.2*10-5).
*Conclusions: Novel computational approaches identify TCR α, β/γ, δ ratios and specific gene expression changes in peripheral blood for non-invasive diagnosis of patients with AMR risk.
Figure 1. Boxplot results of the ANOVA for the classification by transplant outcome using α, β/γ, δ ratio. Vertical axis is percentage of all TCRs which are either TRA or TRB. Results from ANOVA (STA vs. AMR – p-value = 1.0*10-4, STA vs. CMR – p-value = 1.3*10-2).
To cite this abstract in AMA style:
Pineda S, Ravoor A, Bestard O, Sarwal M, Sirota M. Integrative Analysis of Immune Repertoire and Gene Expression from RNA-seq in Kidney Organ Transplants [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/integrative-analysis-of-immune-repertoire-and-gene-expression-from-rna-seq-in-kidney-organ-transplants/. Accessed February 28, 2026.« Back to 2019 American Transplant Congress