Integrative Analyses of Circulating Small Rnas and Paired Kidney Graft Transcriptome in Transplant Glomerulopathy
1Surgery, James D Eason Transplant Institute, UTHSC, Memphis, TN, 2Department of Systems and Computational Biology, School of Life Sciences, University of Hyderabad,, Hyderabad,, India, 3Center for Biomedical Informatics, University of Tennessee Health Science Center, Memphis, TN, 4Surgery, James D Eason Transplant Institute, Memphis, TN, 5Surgery, Program in Transplantation, University of Maryland, Baltimore, MD, 6Surgery, Division Surgical Science, University of Maryland, Baltimore, MD, 7Medicine, Montefiore Medical Center, Abdominal Transplant Program, Albert Einstein College of Medicine,, NYC, NY
Meeting: 2021 American Transplant Congress
Abstract number: 15
Keywords: Gene expression, Genomic markers, Kidney transplantation, Recurrence
Topic: Clinical Science » Biomarkers, Immune Assessment and Clinical Outcomes
Session Information
Session Name: Biomarkers, Immune Assessment and Clinical Outcomes - I
Session Type: Rapid Fire Oral Abstract
Date: Saturday, June 5, 2021
Session Time: 4:30pm-5:30pm
 Presentation Time: 4:50pm-4:55pm
Presentation Time: 4:50pm-4:55pm
Location: Virtual
*Purpose: Transplant glomerulopathy (TG) develops through multiple mechanisms including donor-specific antibodies, T cells and innate immunity. We investigate the role of circulating small RNAs (sRNAs) in TG comparing intragraft gene expression profiles.
*Methods: sRNAs profiles were evaluated in serum of kidney transplant recipients with biopsy-proven TG and normal allograft function using RNA sequencing. An integrative approach was applied to link miRNA changes in serum with gene expression profiles from the paired allograft samples.
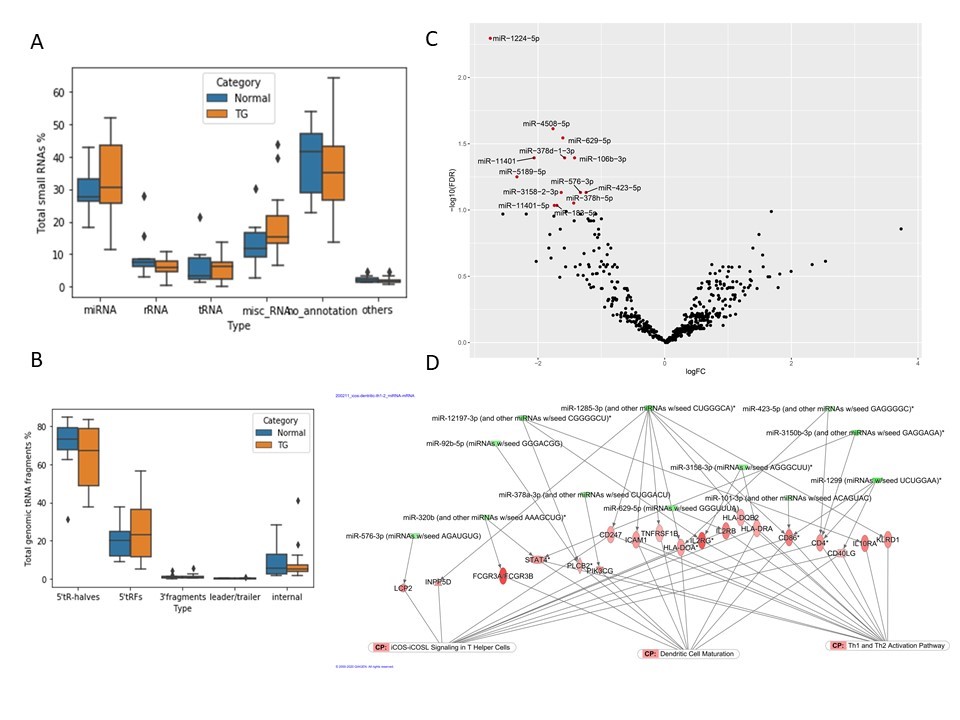
*Results: Among total sRNA population, miRNAs were the most abundant species, compromising approximately 30% of total sRNAs in the serum of kidney transplant patients (Fig. A). Fragments arising from mature tRNA and rRNA were also detected. Most of tRNA fragments (tRFs) were generated from 5’ ends of mature tRNA and mainly from two parental tRNAs: tRNA-Gly and tRNA-Glu (Fig. B). Paired gene expression analysis from allograft tissues showed changes in canonical pathways related to immune activation such as iCos-iCosL signaling pathway in T helper cells, Th1 and Th2 activation pathway, and dendritic cell maturation (Fig. C, D). mRNA targets of down regulated miRNAs such as miR-1224-5p, miR-4508, miR-320, miR-378a from serum were globally upregulated in tissue.
*Conclusions: This is the first study to report the circulating sRNA profiles in transplant patients with TG using next generation sequencing. A novel circulating tRNA fragment TG signature is reported. Integration of sRNA profiles from serum with gene expression from tissues showed that changes in serum miRNAs support the role of T-cell mediated mechanisms in ongoing allograft injury.
To cite this abstract in AMA style:
Kuscu C, Manjari K, Akram M, Kuscu C, Wolen A, Bajwa A, Eason J, Maluf DG, Mas V, Akalin E. Integrative Analyses of Circulating Small Rnas and Paired Kidney Graft Transcriptome in Transplant Glomerulopathy [abstract]. Am J Transplant. 2021; 21 (suppl 3). https://atcmeetingabstracts.com/abstract/integrative-analyses-of-circulating-small-rnas-and-paired-kidney-graft-transcriptome-in-transplant-glomerulopathy/. Accessed February 27, 2026.« Back to 2021 American Transplant Congress