HLA Mismatch Immunogenicity: Are We Predicating Mismatches That Induce De Novo DSA Development or That Do Not?
1Terasaki Research Institute, Los Angeles, CA, 2East Carolina University, Greenville, NC, 3Eastern Nephrology Associates, Greenville, NC
Meeting: 2020 American Transplant Congress
Abstract number: A-286
Keywords: Epitopes, HLA antibodies, Immunogenicity, Kidney transplantation
Session Information
Session Name: Poster Session A: Histocompatibility and Immunogenetics
Session Type: Poster Session
Date: Saturday, May 30, 2020
Session Time: 3:15pm-4:00pm
 Presentation Time: 3:30pm-4:00pm
Presentation Time: 3:30pm-4:00pm
Location: Virtual
*Purpose: Although the development of de novo donor-specific anti-HLA antibodies (dnDSA) remains a major risk factor for poor long-term kidney allograft outcomes, the prediction of dnDSA development has not been fully elucidated. The purpose of this study is to evaluate the potential of machine learning technics to predict dnDSA development.
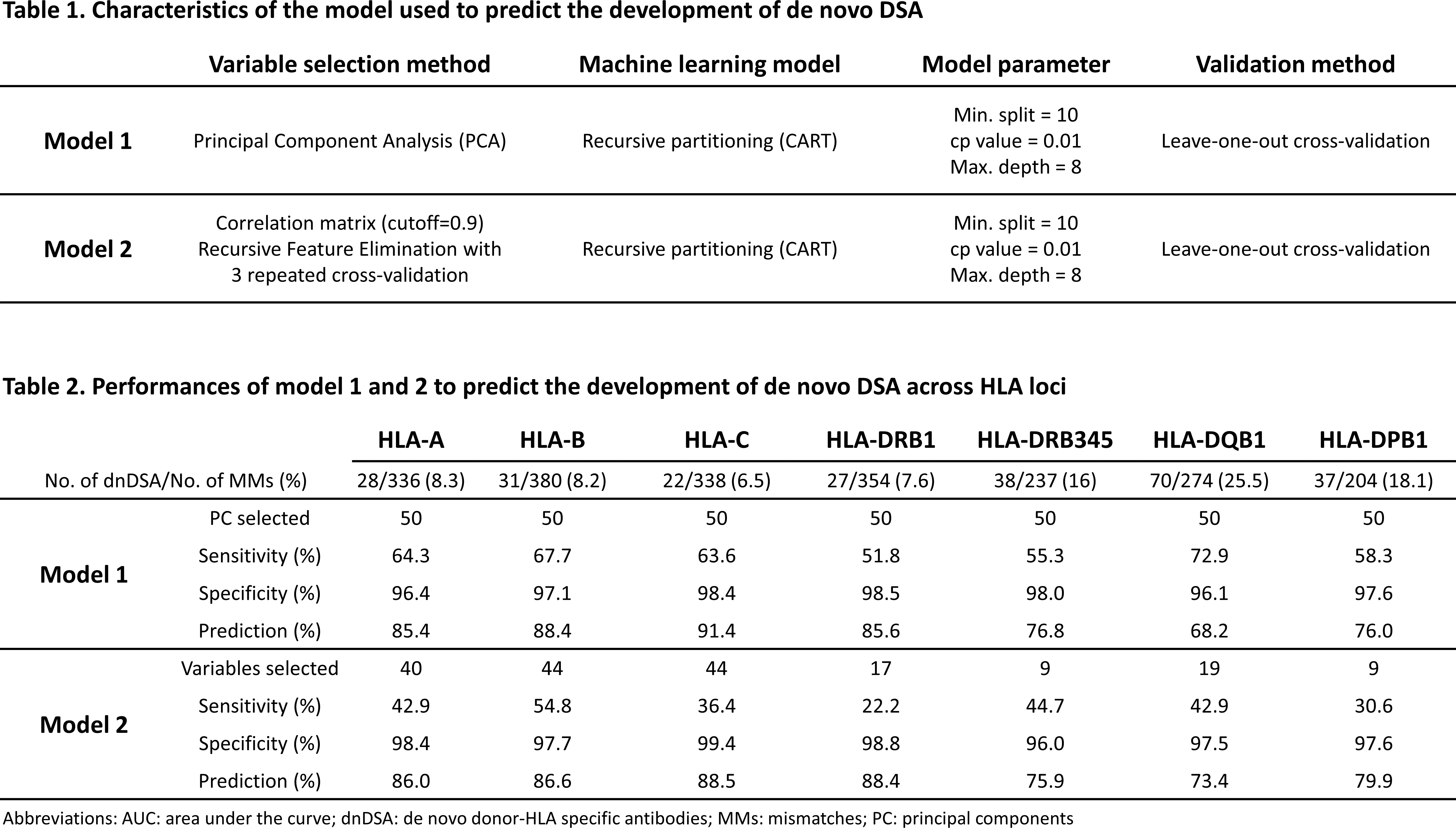
*Methods: We retrospectively studied 236 HLA-mismatched kidney recipients transplanted between 2011 and 2016. All recipients received calcineurin inhibitors, did not have preformed DSA, were HLA-typed by NGS for HLA-A, -B, -C, -DRB1, -DRB345, -DQB1, -DPB1, and were screened for dnDSA using Single Antigen Beads with a median follow-up of 2.26 years. We used E3, an HLA immunogenicity software, to generate 55 molecular features (MoFs) of HLA mismatches (MMs) related to physiochemical difference, HLA antibody epitopes and HLA T cell epitopes. To predict the development of dnDSA against specific HLA MMs, we used a recursive partitioning model with different variable selection methods, and a leave-one-out cross-validation (Table 1).
*Results: Among the 236 recipients, we observed a total of 2123 HLA MMs, of which 328 induced a dnDSA. The number of dnDSA developed against each HLA loci and the performance of model 1 and 2 to predict dnDSA development are listed in Table 2. For model 1 and 2 respectively, the range of: sensitivity was 52-73% and 22-55%; specificity was 96-99% and 96-99%; and prediction was 76-91% and 73-89%; across HLA loci. In both model, the prediction was above 80% for HLA-A-, -B, -C, and -DRB1 MMs, and below 80% for HLA-DRB345, -DQB1, and -DPB1. Last, the sensitivity of model 1 was higher than model 2 overall.
*Conclusions: This study shows that development of dnDSA can be predicted accurately; however, the prediction only reveals which HLA MMs may not induce a dnDSA rather than which HLA MM will induce a dnDSA. Nonetheless, it remains important – from a clinical perspective – to know if certain HLA MMs have a low risk of inducing a dnDSA.
To cite this abstract in AMA style:
Jucaud V, Rebellato L, Briley K, Haisch C, Kendrick S, Jones H, Mclawhorn K, Leeser D, Everly M. HLA Mismatch Immunogenicity: Are We Predicating Mismatches That Induce De Novo DSA Development or That Do Not? [abstract]. Am J Transplant. 2020; 20 (suppl 3). https://atcmeetingabstracts.com/abstract/hla-mismatch-immunogenicity-are-we-predicating-mismatches-that-induce-de-novo-dsa-development-or-that-do-not/. Accessed March 8, 2026.« Back to 2020 American Transplant Congress