Genome-Wide Profiling of BK Polyomavirus Integration in Urothelial Carcinoma of Kidney Transplant Recipients Reveals a Potential Microhomology or Nonhomologous End Joining Mediated Integration Mechanism
1Nanfang Hospital, Southern Medical University, Guangdzhou, China, 2Departments of Medicine, Massachusetts General Hospital, Harvard Medical School, Boston, MA, 3Departments of Urology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, 4Departments of Surgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA, 5Mygenostics Co., Beijing, China, 6Departments of Pathology, Massachusetts General Hospital, Harvard Medical School, Boston, MA
Meeting: 2019 American Transplant Congress
Abstract number: D337
Keywords: Immunosuppression, Kidney transplantation, Malignancy, Polyma virus
Session Information
Session Name: Poster Session D: PTLD/Malignancies: All Topics
Session Type: Poster Session
Date: Tuesday, June 4, 2019
Session Time: 6:00pm-7:00pm
 Presentation Time: 6:00pm-7:00pm
Presentation Time: 6:00pm-7:00pm
Location: Hall C & D
*Purpose: Chronic BK polyomavirus (BKV) infection is recognized as a potential oncogenic factor of urothelial carcinoma (UC) in renal transplant recipients. Besides, a positive correlation among BKV integration, overexpression of viral large T antigen (TAg) and malignancy was reported in recent studies. However, little is known about the integration mechanism; and it remains unclear whether viral integration has any impact on deregulation of viral oncogene expression, host gene expression, and host genomic instability.
*Methods: In this study, whole-genome sequencing and capture-based sequencing were performed on two high-grade urothelial carcinomas with positive TAg expression in two individual renal transplant recipients.
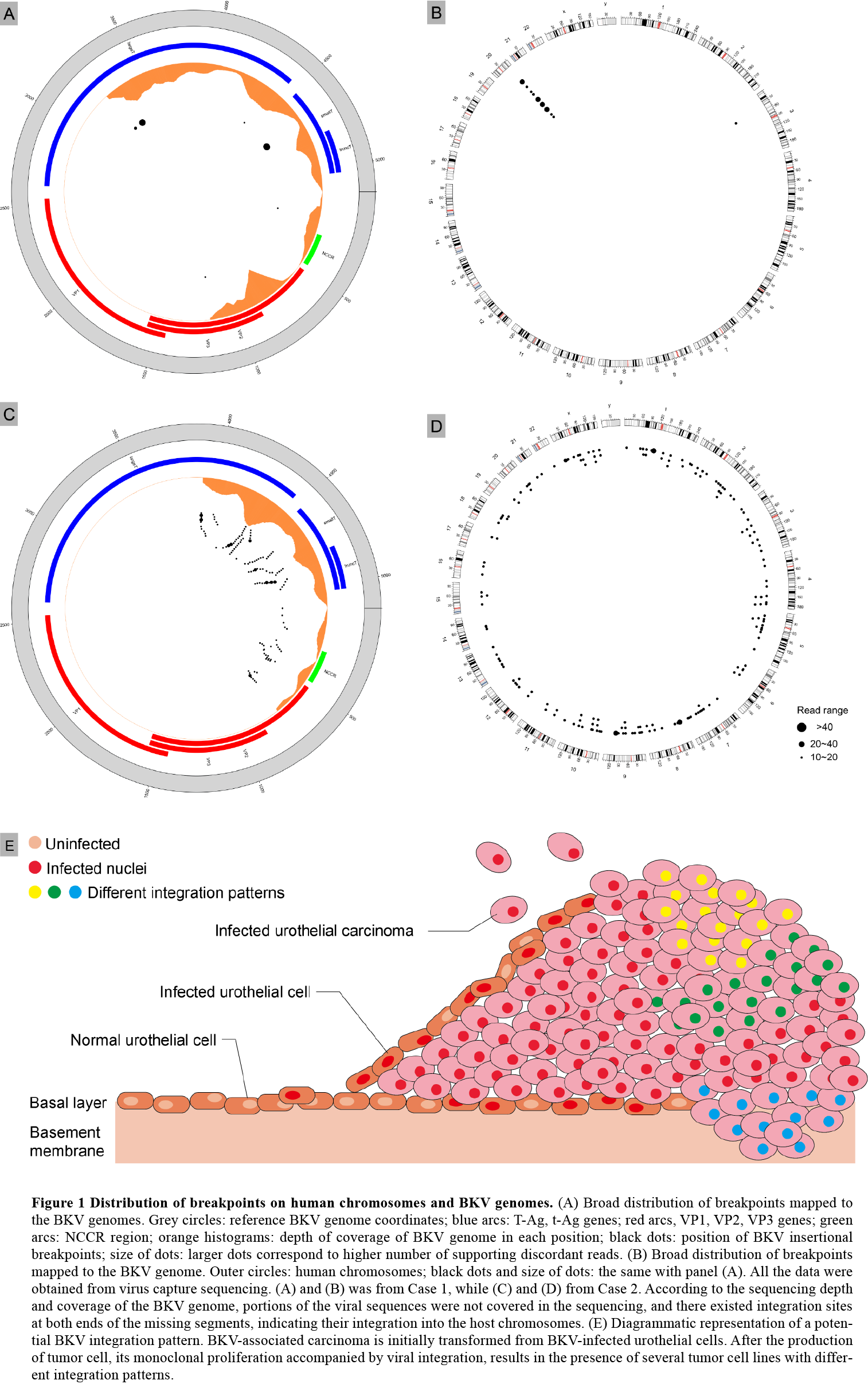
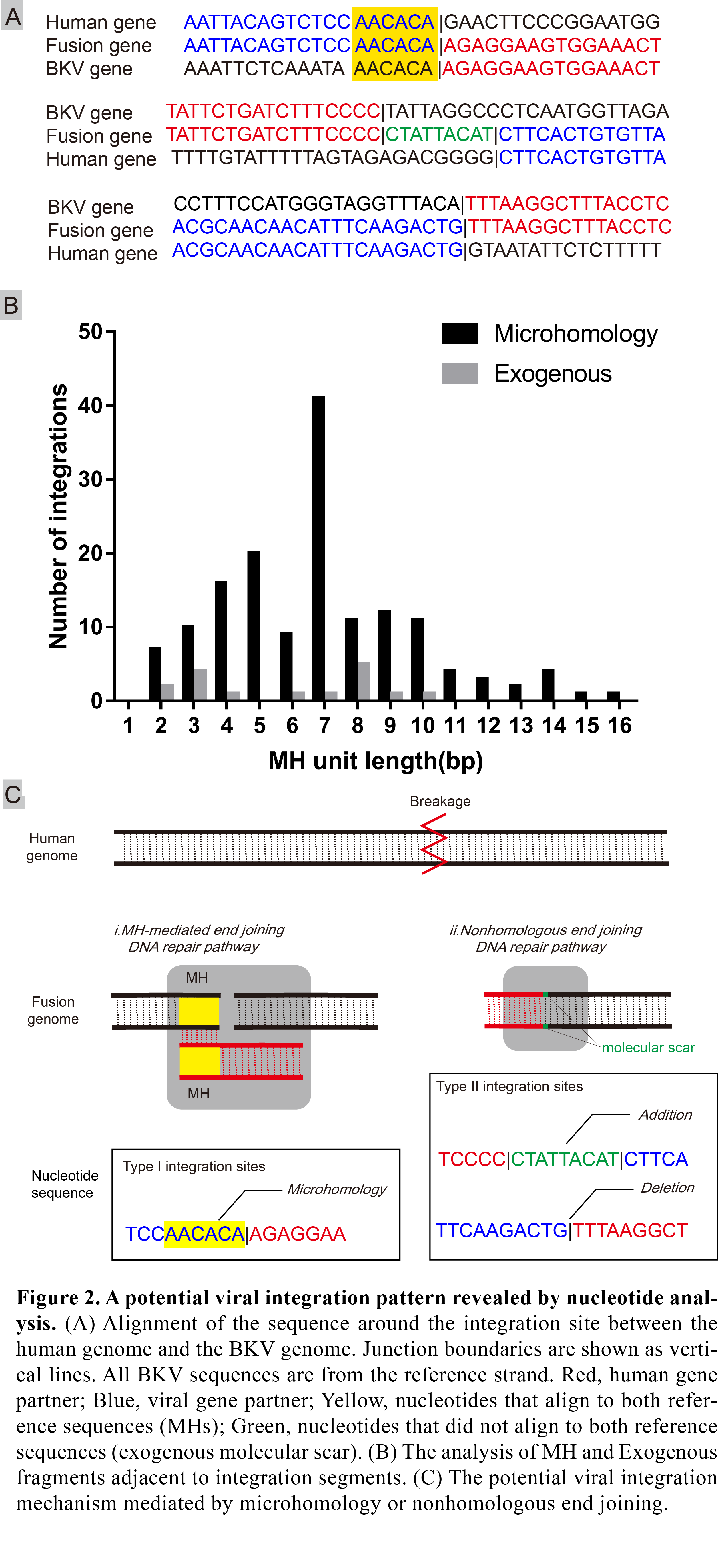
*Results: A total of 185 integration sites from BKV genotype Ic and Ib were identified locating at almost all chromosomes in both cases. The microhomologies (MHs) between the human and the BKV genome were significantly enriched in flanking regions at 84.4% integration sites (157 of 185), with a median of 7 bp for the MHs (range 2,16). 76 human genes were formed fusion sequence due to viral insertional integration, of which 25 genes were statistically associated with urothelial carcinoma using GEO2R analysis.
*Conclusions: Our results indicated a multi-site and multi-fragment linear integration pattern and a potential microhomology or nonhomologous end joining integration mechanism at a single-nucleotide level. Based on these findings we tentatively put forward a selection mechanism driven by immunity and centered on viral integration in the carcinogenesis of BKV.
To cite this abstract in AMA style:
Jin Y, Zhou Y, Deng W, Lee R, Liu Y, Blute M, Liu R, Elias N, Fu F, Guan B, Geng J, Ma J, Zhou J, Liu N, Wu C, Miao Y. Genome-Wide Profiling of BK Polyomavirus Integration in Urothelial Carcinoma of Kidney Transplant Recipients Reveals a Potential Microhomology or Nonhomologous End Joining Mediated Integration Mechanism [abstract]. Am J Transplant. 2019; 19 (suppl 3). https://atcmeetingabstracts.com/abstract/genome-wide-profiling-of-bk-polyomavirus-integration-in-urothelial-carcinoma-of-kidney-transplant-recipients-reveals-a-potential-microhomology-or-nonhomologous-end-joining-mediated-integration-mechani/. Accessed February 26, 2026.« Back to 2019 American Transplant Congress